贝类作为一种非常古老的类群,在进化过程中,颜色演变得丰富多彩。贝类壳色与贝类的警戒伪装、免疫防御及环境适应等具有密切的关系[1]。此外,贝类壳色作为一个可遗传性状,还与贝类的生长、存活和抗逆等表型性状有关,具有重要的育种价值[2]。因此,开展贝类壳色形成机制研究,将有助于更好地理解贝类壳色变异的生态及进化意义,并为壳色性状的遗传育种研究提供借鉴。
随着高通量测序技术的发展,基于组学技术的壳色形成分子机制研究已在多种贝类中开展,尤其在转录组层面开展了大量研究,鉴定获得了许多与贝类壳色形成相关的候选基因或分子通路[3-5]。近年来,蛋白质组学已成为探索生物生命活动规律和分子机制的重要技术,可在大规模水平上研究蛋白质的特征,由此获得蛋白质水平上对相关生物学问题整体而全面的认识[6-7]。以往大多数海洋无脊椎动物的蛋白质组学研究中多依赖二维凝胶电泳技术,但该技术通量较低、重复性较差[8-9]。近年来,以串联质量标签(TMT,tandem mass tags,串联质量标签)和同位素标记相对和绝对定量(iTRAQ)技术为代表的定量蛋白质组学技术,具有通量高、重复性好、准确性高、数据丰富和自动化程度化高等优点,在蛋白质组学领域得到广泛运用。目前,蛋白质组学技术在贝类中已得到了一定应用,涉及贝类的生长、发育、免疫、神经调控和环境适应等[10-13],但在贝类壳色形成机制研究中应用还相对较少,限制了对贝类壳色形成分子机制的深入理解。
虾夷扇贝(Patinopecten yessoensis)是中国北方特色冷水性经济贝类,原产于日本、朝鲜和俄罗斯,体型较大,营养丰富,具有重要的经济价值[14]。虾夷扇贝壳色呈现简单的多态性(图1),是研究壳色较好的材料。常见的虾夷扇贝,右壳较突,呈白色,左壳稍平,呈褐色;群体中少数个体左右壳皆为白色。Ding等[15]对不同壳色品系虾夷扇贝外套膜进行转录组测序发现,甜菜红色素生物合成及酪氨酸代谢过程在虾夷扇贝壳色形成中可能发挥重要作用。Mao等[16]对虾夷扇贝外套膜不同区域的转录组测序发现,黑色素合成过程在差异表达基因中显著富集,很可能与壳色形成有关。Yuan等[17]对不同壳色虾夷扇贝外套膜的全基因组DNA甲基化测序发现,黑色素和血红色合成通路基因甲基化水平在不同壳色间存在显著差异,表明虾夷扇贝壳色形成过程受DNA甲基化的调控。然而,目前在蛋白质组学层面上的虾夷扇贝壳色形成机制研究还未见报道。
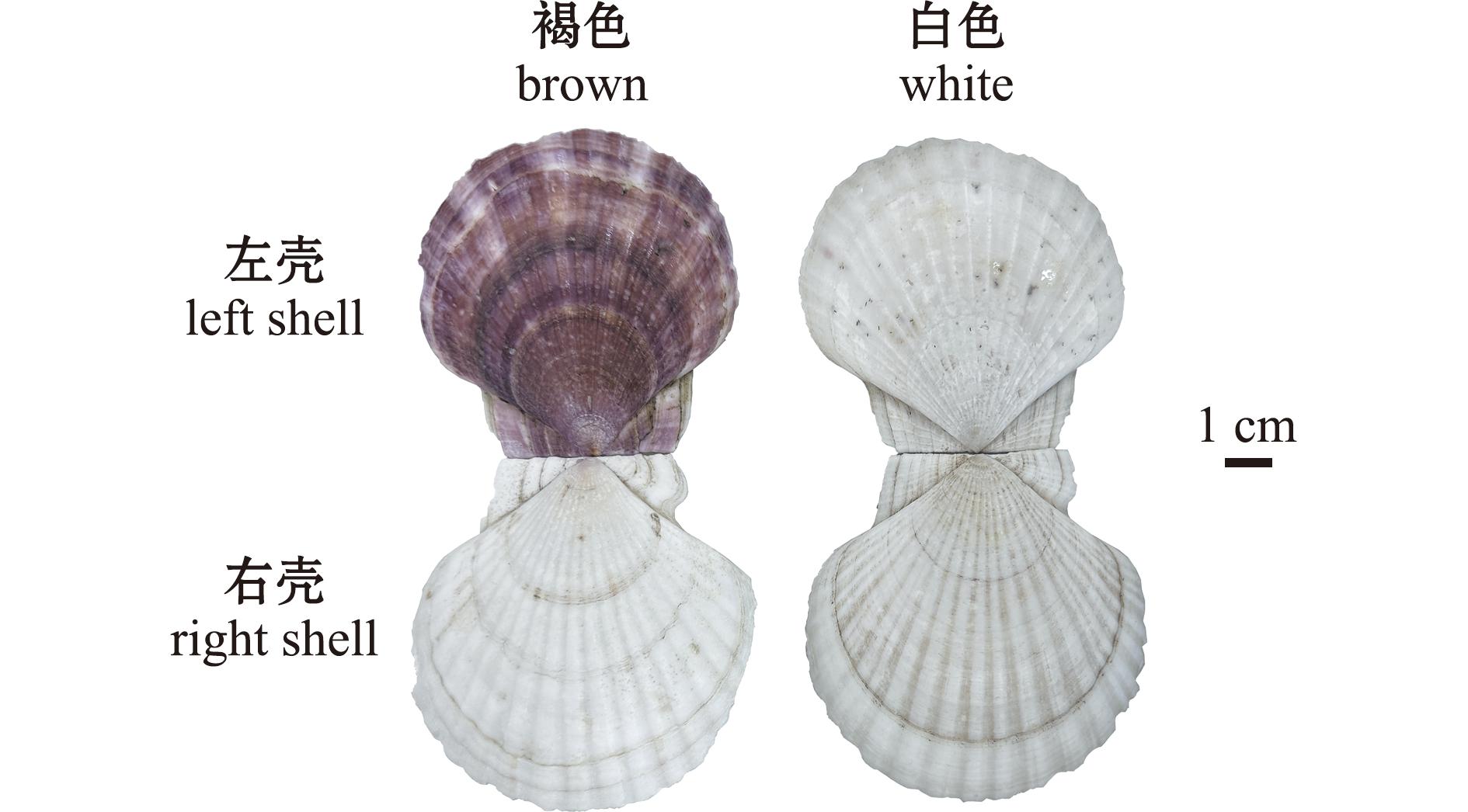
图1 两种壳色虾夷扇贝贝壳
Fig.1 Shells of Yesso scallops with two shell colours
本研究中以虾夷扇贝为材料,利用TMT标记定量技术进行褐色和白色两种壳色虾夷扇贝外套膜的蛋白质组学分析,筛选两种壳色虾夷扇贝间的差异表达蛋白并进行功能富集及蛋白互作分析,挖掘与壳色形成相关的蛋白及分子通路,以期从蛋白质组水平研究虾夷扇贝壳色形成的分子调控机制。
1 材料与方法
1.1 材料
试验所用2龄健康褐色和白色两种壳色虾夷扇贝(图1)采自大连獐子岛海域底播群体。在实验室条件下进行一周暂养,过滤海水充气并维持水温在10 ℃左右,每天投喂小球藻(Chlorella sp.)两次,更换海水一次。一周后,每种壳色类型扇贝随机选取活性较好的3只进行解剖,对两种壳色扇贝左侧外套膜组织进行取样(褐色扇贝编号为B1~B3,白色扇贝编号为W1~W3),并立即用液氮冷冻,并于-80 ℃的超低温冰箱中保存,用于TMT标记蛋白质组学分析。
1.2 方法
1.2.1 蛋白质的提取 将外套膜组织在液氮中充分研磨至粉末,移至1.5 mL离心管中,加入酚抽提取液600 μL,并加入苯甲硫酰氟(PMSF)使其终浓度为1 mmol/L,冰上超声破碎;加入等体积酚-Tris-HCl溶液(pH 7.8),4.0 ℃下摇匀混合30 min,7 100 g离心10 min,收集上清液;加入5倍体积预冷0.1 mol/L醋酸铵-甲醇溶液,于-20 ℃过夜沉淀,以12 000 g离心10 min,收集沉淀;加入5倍体积预冷甲醇清洗沉淀,4 ℃下12 000 g离心10 min收集沉淀,重复此过程1次;然后,以丙酮代替甲醇重复上述步骤两次,去除甲醇。将沉淀在室温下干燥约5 min,并溶解于样品裂解液;最后,将溶液12 000 g离心10 min取上清,以充分去除沉淀。采用BCA蛋白浓度测定方法测定蛋白浓度。
1.2.2 胰蛋白酶酶解及肽段标记 每个样品取100 μg的蛋白质,加入二硫苏糖醇(DTT)溶液使其终浓度为4.5 mmol/L,55 ℃下孵育30 min;冰上冷却至室温,加入碘乙酰胺使其终浓度为9 mmol/L,充分混匀,室温避光放置15 min。加入6倍体积冷丙酮,-20 ℃过夜,沉淀蛋白;4 ℃,8 000 g离心10 min,离心收集沉淀,加入100 μL 300 mmol/L四乙基溴化铵(TEAB)溶解沉淀,加入2 μg胰酶Trypsin-TPGK,37 ℃下过夜消化。酶解后的肽段经冷冻干燥后备用。
向冻干样品中加入66 μL 200 mmol/L TEAB缓冲液,混匀,取30 μL样品使用TMT10plexTM试剂盒进行标记。每个样品中加入41 μL TMT试剂进行混合,室温放置1 h,加入8 μL 5%羟胺终止反应,冻干,-80 ℃下保存。各样品标记如下:B1-126,B2-127N,B3-127C,W1-128N,W2-128C,W3-129N。
1.2.3 反向色谱分离 标记后的蛋白样品,上样液相色谱(Agilent 1100 HPLC)进行分离,色谱柱为Agilent Zorbax Extend-C18窄径柱。洗脱液分别为流动相A(2%乙腈,乙腈∶水=2∶98)和流动相B(90%乙腈,乙腈∶水=90∶10)。洗脱梯度设置为0~8 min,98%A;8~8.01 min,98%~95%A;8.01~48 min,95%~75%A;48~60 min,75%~60%A;60~60.01 min,60%~10%A;60.01~70 min,10%A;70~70.01 min,10%~98%A;70.01~75 min,98%A。流速为300 μL/min,紫外检测波长分别为210、280 nm。收集8~60 min内的样品,每分钟收集洗脱液到1~15号离心管中,按顺序收回样品进行真空冷冻干燥,用于LC-MS/MS分析。
1.2.4 LC-MS/MS分析 将样品以300 nL/min流速上样到色谱柱预柱Acclaim PepMap100 (RP-C18,Thermo Fisher),再经分析柱Acclaim PepMap RSLC,(RP-C18,Thermo Fisher)进行分离。洗脱液分别为流动相A (0.1%甲酸,甲酸∶水=0.1∶99.9)和流动相B (80%乙腈,0.1%甲酸,乙腈∶水∶甲酸=80∶19.9∶0.1)。洗脱梯度为0~1 min,2%~9% B;1~45 min,9%~29% B;45~52 min,29%~37% B;52~56 min,37%~100% B;56~60 min,100% B。MS质谱扫描使用350~1 500的荷质比,质量分辨率为60 000,自动增益控制值为3×106;对MS质谱中的10个最高峰进行能碰撞裂解,碰撞能量为32;MS/MS图谱采集的分辨率为45 000,自动增益控制为2×105,离子最大注射时间为80 ms,动态排除时间为30 s。本试验获得的原始质谱数据已提交至蛋白质组学数据库iProX(https://www.iprox.cn/),数据编号为IPX0004367000。
1.2.5 蛋白质鉴定及生物信息学分析 利用Proteome Discover 2.4搜索虾夷扇贝基因组数据库[18],进行虾夷扇贝外套膜组织蛋白质的鉴定(Score Sequest HT>0、unique peptide≥2和FDR<1%)。随后,以P value<0.05,Foldchange≥1.2为阈值进行差异表达蛋白(differentially expressed proteins,DEPs)的筛选。利用GO(Gene Ontology)数据库[19]和KEGG (Kyoto Encyclopedia of Genes and Genomes)数据库[20]进行差异表达蛋白的功能富集功能,以gene number≥2,q value<0.05作为显著富集的阈值。利用String 数据库(https://string.embl.de/)进行差异蛋白互作网络分析(PPi),采用Cytoscape软件(https://www.cytoscape.org/)绘图。
1.2.6 实时荧光定量PCR 实时荧光定量PCR(quantitative real-time PCR,qRT-PCR)试验参考杭雲娜等[21]的方法。简要概括如下:利用RNAprep Pure 动物组织总 RNA 提取试剂盒(Tiangen)进行褐色和白色虾夷扇贝左侧外套膜总RNA的提取。利用琼脂糖凝胶电泳检测RNA的完整性,利用NV3000 型显微分光光度计(Vastech)测定RNA的纯度和浓度。采用PrimeScript RT reagent试剂盒(TaKaRa)反转录合成第一链cDNA,将所有cDNA产物稀释至200 ng/μL,作为qRT-PCR的模板。采用FastStart Essential DNA Green Master试剂盒(Roche)在Light Cycler 96 System荧光定量PCR仪(Roche)上进行反应。20 μL反应体系包括:上游引物(10 μmol/L) 0.8 μL,下游引物(10 μmol/L)0.8 μL,FastStart Essential DNA Green Master 10 μL,ddH2O 6.4 μL,cDNA模板2 μL。反应程序如下:95 ℃下预变性10 min;然后,95 ℃下变性10 s,60 ℃下退火30 s,共进行40次循环;然后,95 ℃下复性10 s,65 ℃下退火1 min;最后再在97 ℃下延伸1 s,37 ℃下退火30 s。以β-actin基因作为内参基因。各基因引物序列见表1。利用Blast软件(BLASTN)将引物序列与虾夷扇贝基因组和转录组进行比对检测引物特异性(E-value≤l×10-10),并对反应产物进行熔解曲线分析,保证单个扩增产物。每个反应进行3个技术重复,每种壳色扇贝进行3个生物学重复。采用2-ΔΔCt方法进行两种壳色虾夷扇贝DEP mRNA表达水平的分析。
表1 实时荧光定量PCR引物序列
Tab.1 Primer sequences used for qRT-PCR

引物primer登录号accessionnumber引物序列(5′-3′)primersequence(5′-3′)TKTA0A210PYJ0F:AACAACCAGTCAGCCACAR:GAAAGCCGTCATCCAACACAM-A0A210PKQ2F:ATCATCTCCCGCCTCACTR:TCCATCAGCGTCCACCTCPSOTNA0A210QAG1F:CGACGAGGTGCTTAGTGTR:GGGAATAGCTGGATTTGATTKA0A210PXY8F:GTATGTTGCTGGTGGTGCR:ATTTGTTTGAGCCCTTCTHEBP2A0A210Q959F:AGACTTCCCAGGATTTGCR:TCACTTAGCCACGAACCAPYA0A210QQN6F:ATTGGAGAAGCGAGAACCR:ACAATCACCACCAACCCTCYP3A24A0A210QUD3F:ACGGAAGAAGAAATAACGR:CCTCAACATAGGTGGAAAC1QTNF6A0A210QDK9F:CAGGATGTGATGAGGGTTR:GTCGCTAACAAATGTCCACTRGNA0A210QK55F:CCGCAAGTACCGAACGACR:TATCAACGAAGACGAGGCCOL6A5A0A210QLL6F:ACGGAGACCCAGTTGATAR:TCGTCGCTGTAGGTGATACYSA0A210QHS7F:GCATTAAGCAGGTCGTACR:GACTCAATCGGATCGTTGAMcaseA0A210QEH0F:TGCTGGCTGTGGGAGGATR:TTGGCTTTGTCTGAGGTTGLUA0A210PPN6F:AAACGACCGAGCATTGACR:TCGCAGGTTGGACTCTTGLIPA0A210QHG1F:GGCAGGACTCAATTACGCR:GACCATTTCAGCCCACTAβ-actin—F:CCAAAGCCAACAGGGAAAAGR:TAGATGGGGACGGTGTGAGTG
1.3 数据处理
采用SPSS软件独立T样本检验法进行两种壳色扇贝基因表达水平的差异分析,显著性差异设为0.05。
2 结果与分析
2.1 虾夷扇贝外套膜蛋白质的鉴定
本研究中利用TMT标记定量法对两种壳色虾夷扇贝的6个外套膜组织样品(每组设置3个生物学重复)进行蛋白质组学分析。经过鉴定,共获得433 319个二级图谱,64 317个有效图谱,获得肽段33 882个,可信蛋白质5 177个。对鉴定获得的蛋白质进行统计发现,蛋白质分子质量主要集中在0~250 000(图2(a)),每个蛋白对应的肽段数主要在0~25(图2(b)),各肽段相对于完整蛋白序列的覆盖度主要集中在0%~20% (图2(c))。对各蛋白质的表达量进行主成分分析,发现同一壳色组的样品在空间上分布比较集中(图2(d)),表明各组样品的重复性较好。
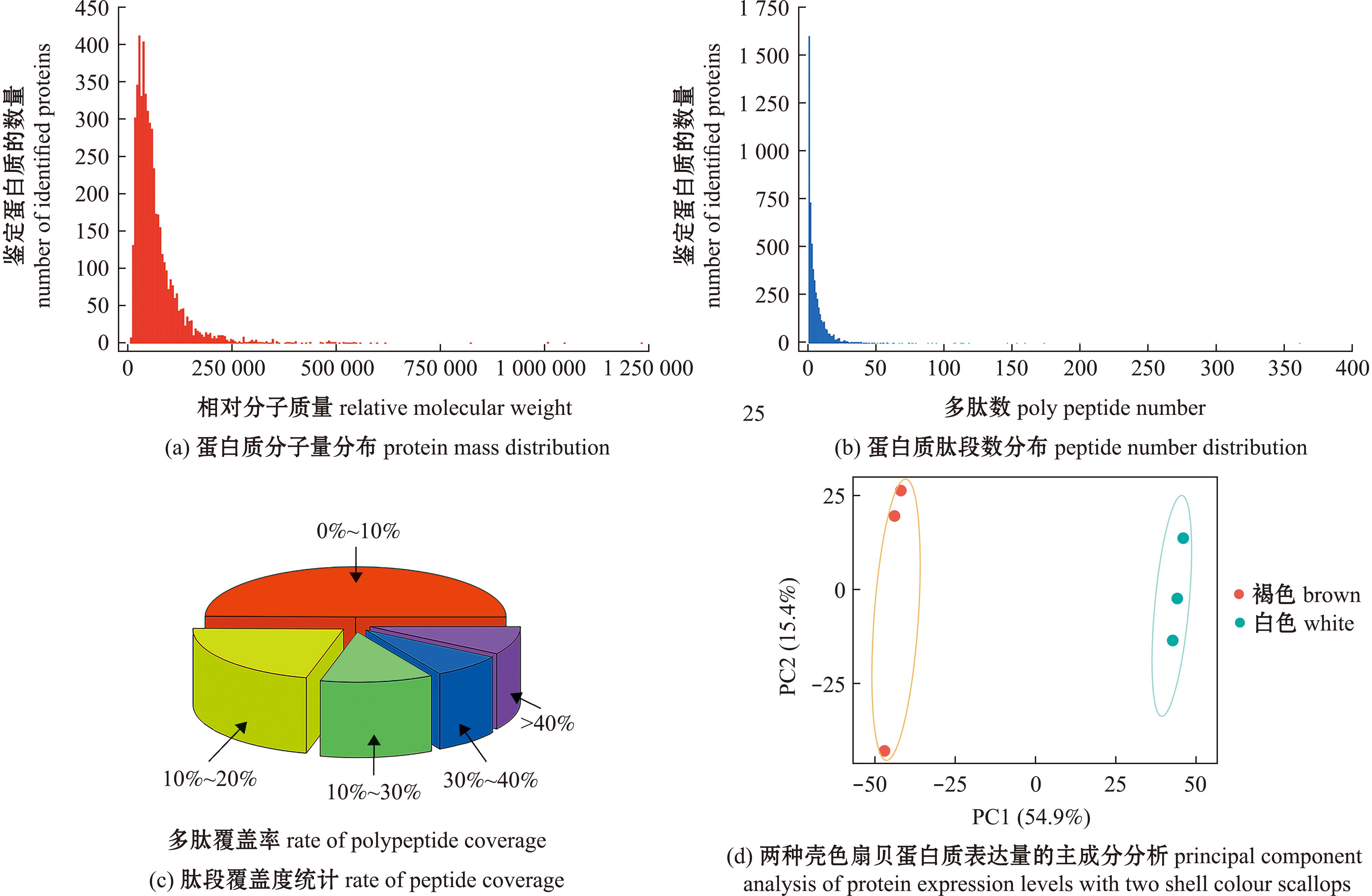
图2 两种壳色虾夷扇贝蛋白质组分析基本信息
Fig.2 Basic information on proteomic analysis of Yesso scallops with two shell colours
2.2 不同壳色虾夷扇贝间差异表达蛋白的筛选
总共筛选获得343个蛋白在褐色与白色虾夷扇贝外套膜中差异表达。其中,187个DEPs在白色虾夷扇贝外套膜中上调表达,156个DEPs下调表达(图3(a)、(b))。两种壳色虾夷扇贝间DEPs表达水平聚类分析表明,两种壳色扇贝的蛋白表达间存在明显差异(图3(c))。这些差异表达蛋白中有许多与壳或壳色的形成、免疫、神经调控及DNA甲基化相关(表2)。
表2 两种壳色虾夷扇贝中的重要候选差异表达蛋白
Tab.2 Important candidate DEPs from Yesso scallops with two shell colour
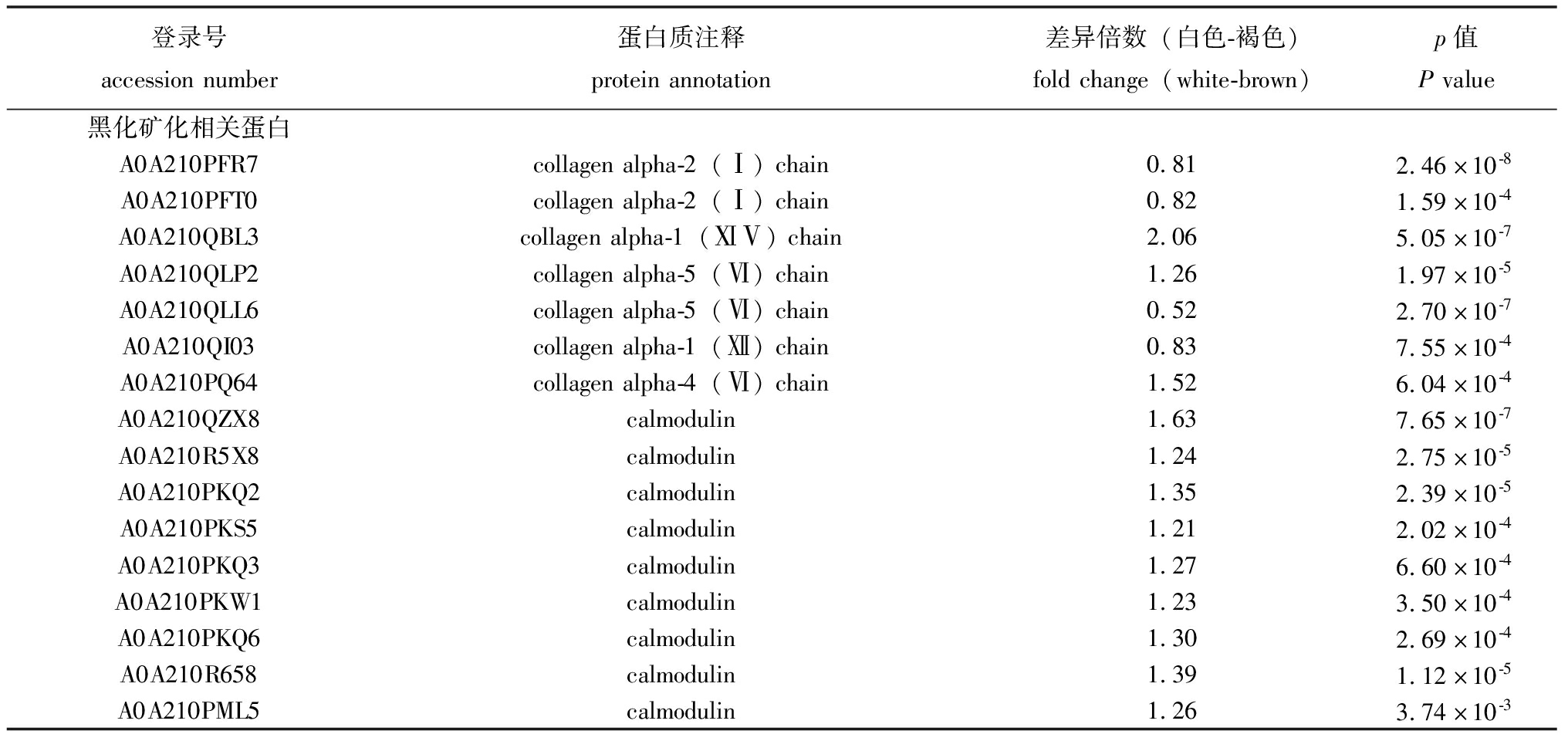
登录号accessionnumber蛋白质注释proteinannotation差异倍数(白色-褐色)foldchange(white-brown)p值Pvalue黑化矿化相关蛋白A0A210PFR7collagenalpha-2(Ⅰ)chain0.812.46×10-8A0A210PFT0collagenalpha-2(Ⅰ)chain0.821.59×10-4A0A210QBL3collagenalpha-1(ⅪⅤ)chain2.065.05×10-7A0A210QLP2collagenalpha-5(Ⅵ)chain1.261.97×10-5A0A210QLL6collagenalpha-5(Ⅵ)chain0.522.70×10-7A0A210QI03collagenalpha-1(Ⅻ)chain0.837.55×10-4A0A210PQ64collagenalpha-4(Ⅵ)chain1.526.04×10-4A0A210QZX8calmodulin1.637.65×10-7A0A210R5X8calmodulin1.242.75×10-5A0A210PKQ2calmodulin1.352.39×10-5A0A210PKS5calmodulin1.212.02×10-4A0A210PKQ3calmodulin1.276.60×10-4A0A210PKW1calmodulin1.233.50×10-4A0A210PKQ6calmodulin1.302.69×10-4A0A210R658calmodulin1.391.12×10-5A0A210PML5calmodulin1.263.74×10-3
续表2 两种壳色虾夷扇贝中的重要候选差异表达蛋白
Cont.Tab.2 Important candidate DEPs from Yesso scallops with two shell colour
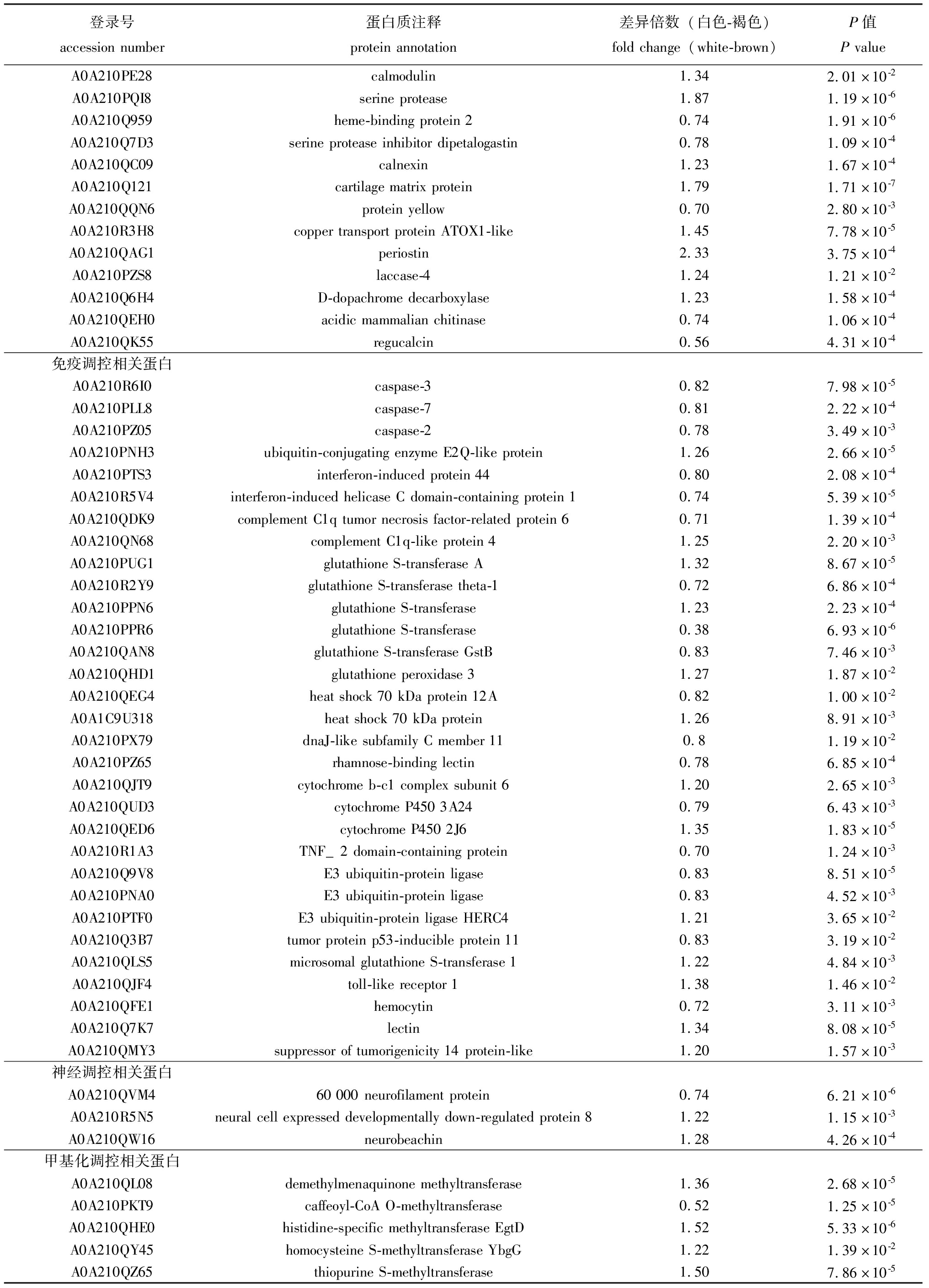
登录号accessionnumber蛋白质注释proteinannotation差异倍数(白色-褐色)foldchange(white-brown)P值PvalueA0A210PE28calmodulin1.342.01×10-2A0A210PQI8serineprotease1.871.19×10-6A0A210Q959heme-bindingprotein20.741.91×10-6A0A210Q7D3serineproteaseinhibitordipetalogastin0.781.09×10-4A0A210QC09calnexin1.231.67×10-4A0A210Q121cartilagematrixprotein1.791.71×10-7A0A210QQN6proteinyellow0.702.80×10-3A0A210R3H8coppertransportproteinATOX1-like1.457.78×10-5A0A210QAG1periostin2.333.75×10-4A0A210PZS8laccase-41.241.21×10-2A0A210Q6H4D-dopachromedecarboxylase1.231.58×10-4A0A210QEH0acidicmammalianchitinase0.741.06×10-4A0A210QK55regucalcin0.564.31×10-4免疫调控相关蛋白A0A210R6I0caspase-30.827.98×10-5A0A210PLL8caspase-70.812.22×10-4A0A210PZ05caspase-20.783.49×10-3A0A210PNH3ubiquitin-conjugatingenzymeE2Q-likeprotein1.262.66×10-5A0A210PTS3interferon-inducedprotein440.802.08×10-4A0A210R5V4interferon-inducedhelicaseCdomain-containingprotein10.745.39×10-5A0A210QDK9complementC1qtumornecrosisfactor-relatedprotein60.711.39×10-4A0A210QN68complementC1q-likeprotein41.252.20×10-3A0A210PUG1glutathioneS-transferaseA1.328.67×10-5A0A210R2Y9glutathioneS-transferasetheta-10.726.86×10-4A0A210PPN6glutathioneS-transferase1.232.23×10-4A0A210PPR6glutathioneS-transferase0.386.93×10-6A0A210QAN8glutathioneS-transferaseGstB0.837.46×10-3A0A210QHD1glutathioneperoxidase31.271.87×10-2A0A210QEG4heatshock70kDaprotein12A0.821.00×10-2A0A1C9U318heatshock70kDaprotein1.268.91×10-3A0A210PX79dnaJ-likesubfamilyCmember110.81.19×10-2A0A210PZ65rhamnose-bindinglectin0.786.85×10-4A0A210QJT9cytochromeb-c1complexsubunit61.202.65×10-3A0A210QUD3cytochromeP4503A240.796.43×10-3A0A210QED6cytochromeP4502J61.351.83×10-5A0A210R1A3TNF_2domain-containingprotein0.701.24×10-3A0A210Q9V8E3ubiquitin-proteinligase0.838.51×10-5A0A210PNA0E3ubiquitin-proteinligase0.834.52×10-3A0A210PTF0E3ubiquitin-proteinligaseHERC41.213.65×10-2A0A210Q3B7tumorproteinp53-inducibleprotein110.833.19×10-2A0A210QLS5microsomalglutathioneS-transferase11.224.84×10-3A0A210QJF4toll-likereceptor11.381.46×10-2A0A210QFE1hemocytin0.723.11×10-3A0A210Q7K7lectin1.348.08×10-5A0A210QMY3suppressoroftumorigenicity14protein-like1.201.57×10-3神经调控相关蛋白A0A210QVM460000neurofilamentprotein0.746.21×10-6A0A210R5N5neuralcellexpresseddevelopmentallydown-regulatedprotein81.221.15×10-3A0A210QW16neurobeachin1.284.26×10-4甲基化调控相关蛋白A0A210QL08demethylmenaquinonemethyltransferase1.362.68×10-5A0A210PKT9caffeoyl-CoAO-methyltransferase0.521.25×10-5A0A210QHE0histidine-specificmethyltransferaseEgtD1.525.33×10-6A0A210QY45homocysteineS-methyltransferaseYbgG1.221.39×10-2A0A210QZ65thiopurineS-methyltransferase1.507.86×10-5
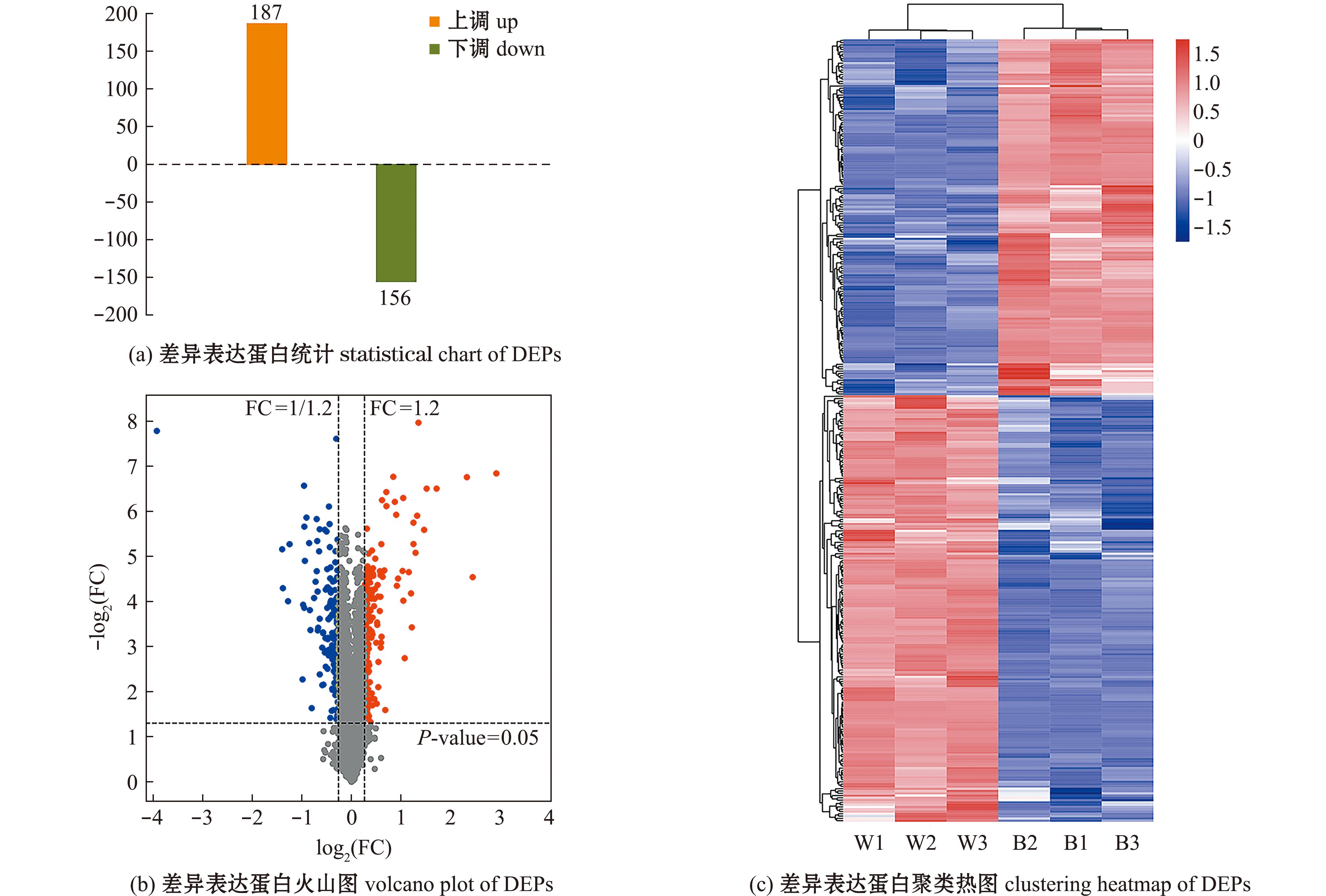
图3 两种壳色虾夷扇贝外套膜的蛋白质差异表达分析
Fig.3 Differential expression analysis of proteins in mantle tissues from Yesso scallops with two shell colour
2.3 差异表达蛋白的GO富集分析
为进一步确定对两种壳色扇贝间DEPs的功能,对其进行GO富集分析。以gene number≥2和q value≤0.05为阈值,共获得161个显著富集的GO条目,其中,97个属于生物学过程(biological process),18个属于细胞组分(cellular component),46个属于分子功能(molecular function)。值得注意的是,这些显著富集的GO条目中,许多与壳色形成和生物矿化、免疫及神经调控有关(图4)。如与壳色形成和生物矿化相关的GO条目有黑色素生物合成过程(melanin biosynthetic process,GO:0042438)、Wnt-蛋白结合(Wnt-protein binding,GO:0017147)、Wnt信号通路的负调控(negative regulation of Wnt signaling pathway,GO:0030178)、胶原三聚体(collagen trimer,GO:0005581)、骨化的负调控(negative regulation of ossification,GO:0030279)等;与免疫相关的GO条目有免疫系统过程(immune system process,GO:0002376)、先天免疫应答(innate immune response,GO:0045087)、先天免疫应答调节(GO:0045088,regulation of innate immune response)、谷胱甘肽转移酶活性(GO:0004364,glutathione transferase activity)等;与神经调控有关的GO条目有乙酰胆碱脂酶活性(acetylcholinesterase activity,GO:0003990)、突触膜(synaptic membrane,GO:0097060)、轴突生成的负调控(negative regulation of axonogenesis,GO:0050771)等。
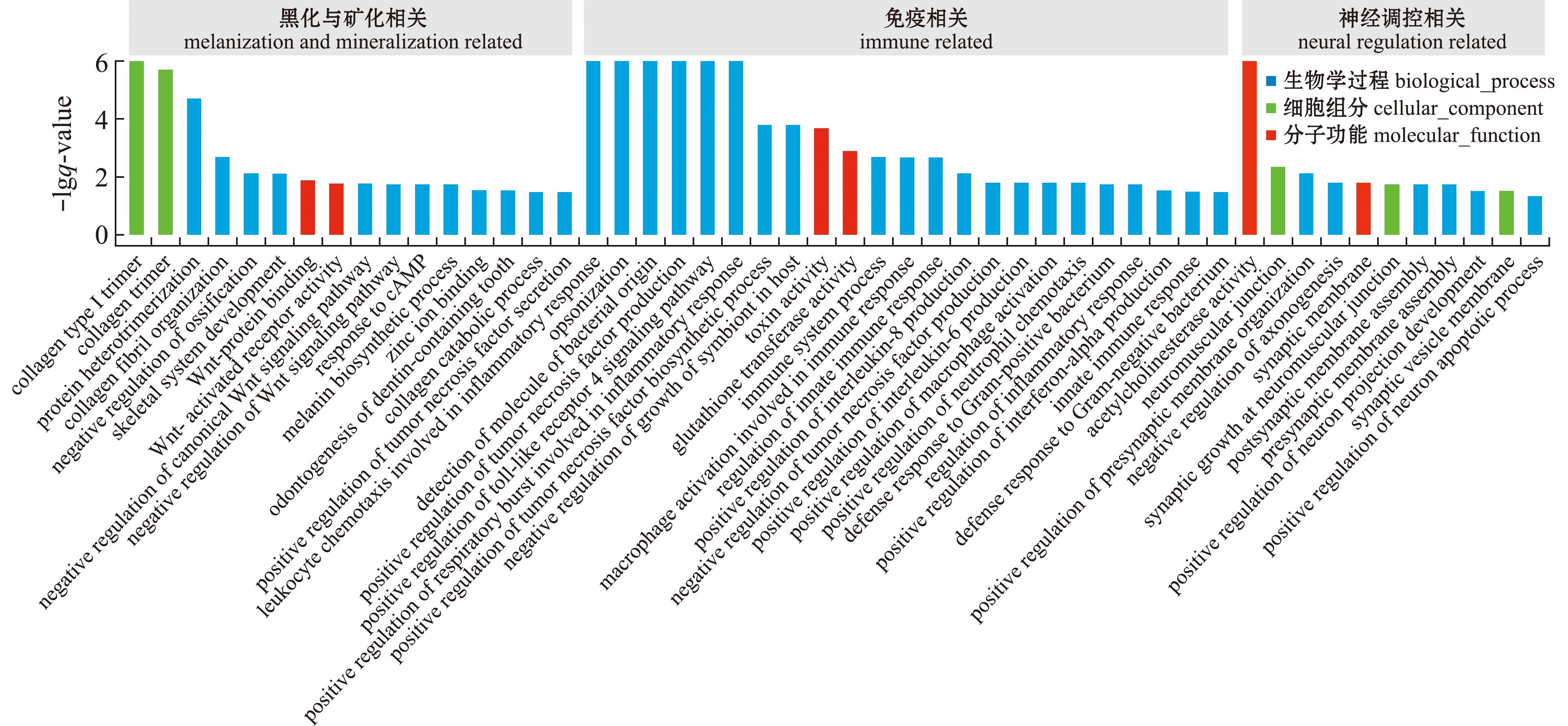
图4 两种壳色虾夷扇贝差异表达蛋白GO富集分析
Fig.4 GO enrichment analysis for DEPs from Yesso scallops with two shell colour
2.4 差异表达蛋白的KEGG富集分析
对两种壳色扇贝间的DEPs进行KEGG富集分析以确定DEPs参与的分子通路。在gene number≥2和q value≤0.05的阈值下,共获得41条显著富集的KEGG通路(图5),包括与壳色形成相关的黑色素生成通路(melanogenesis,ko04024)、钙离子信号通路(calcium signaling pathway,ko04020)、cAMP信号通路(cAMP signaling pathway;ko04024)等。此外一些与免疫相关的分子通路,如TRP通道的炎症介质调节(inflammatory mediator regulation of TRP channels,ko04750)、药物代谢-细胞色素P450(drug metabolism-cytochrome P450)、谷胱甘肽代谢(glutathione metabolism)等,以及与神经调控相关的分子通路,如多巴胺能突触(dopaminergic synapse,ko04728)、神经营养因子信号通路(neurotrophin signaling pathway,ko04722)等,也均显著富集。
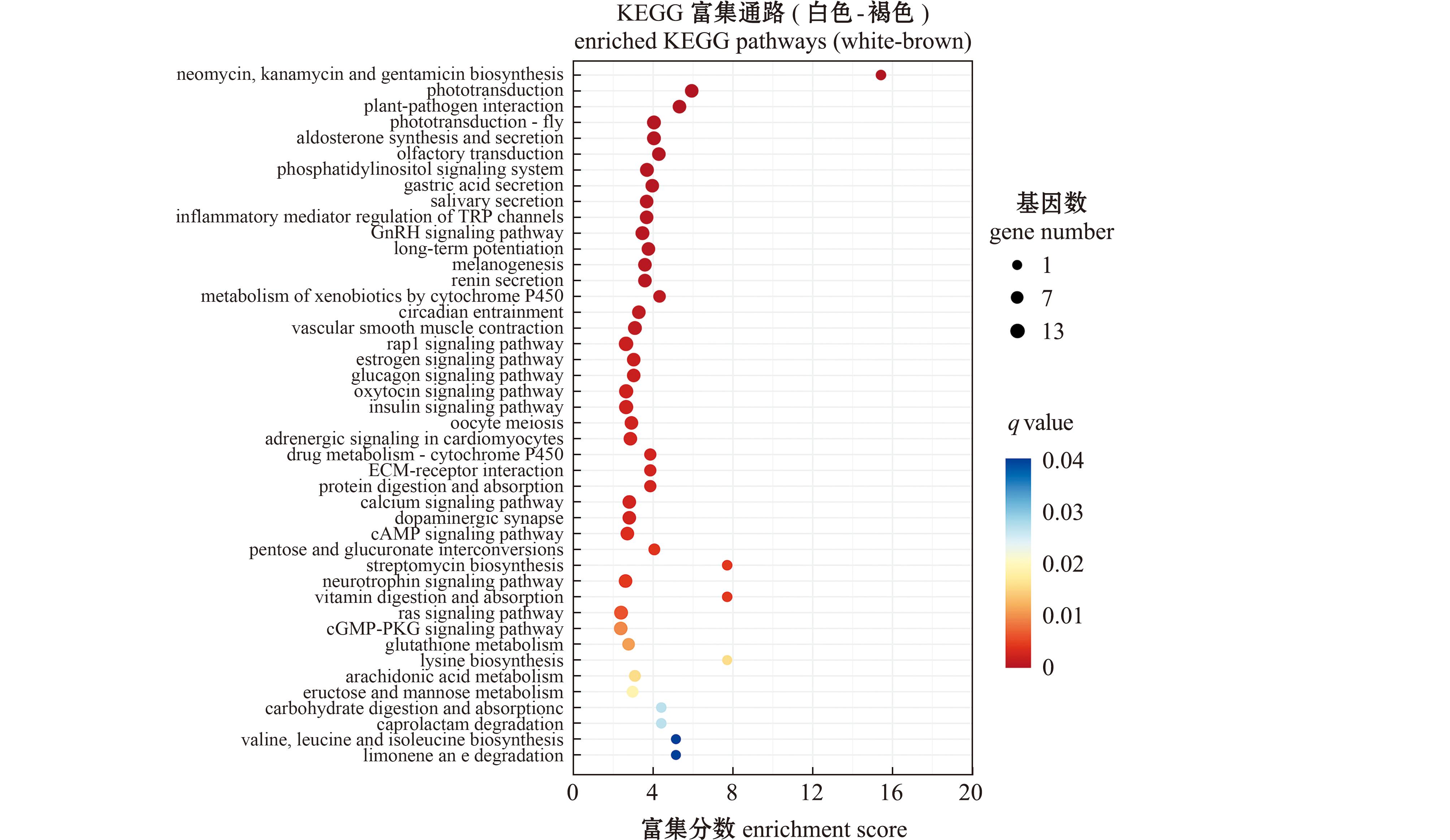
图5 两种壳色虾夷扇贝差异表达蛋白KEGG富集分析
Fig.5 KEGG enrichment for DEPs from Yesso scallops with two shell colour
2.5 差异蛋白互作网络分析(PPi)
利用String 数据库预测了两种壳色虾夷扇贝差异表达蛋白间的互作关系,利用Cytoscape软件构建了差异表达蛋白互作网络图。从图6可见,共预测获得320条互作关系对,互作网络中包含了与壳色和生物矿化相关的蛋白,如Calmodulin(A0A210R658)、Collagen(A0A210QLL6)、Calnexin(A0A210QC09)、Serine protease(A0A210PQI8)、Chitinase(A0A210QEH0)等。此外,网络中还发现一些与免疫、神经及甲基化调控相关的蛋白,如免疫调控相关蛋白Caspase-3(A0A210R6I0)、Caspase-7(A0A210PLL8),神经调控相关蛋白 60 000 neurofilament protein(A0A210QVM4)、Neural cell expressed developmentally down-regulated protein 8(A0A210R5N5),甲基化调控相关蛋白Demethylmenaquinone methyltransferase(A0A210QL08)等,这为后续壳色形成相关蛋白的互作机制研究提供了依据。
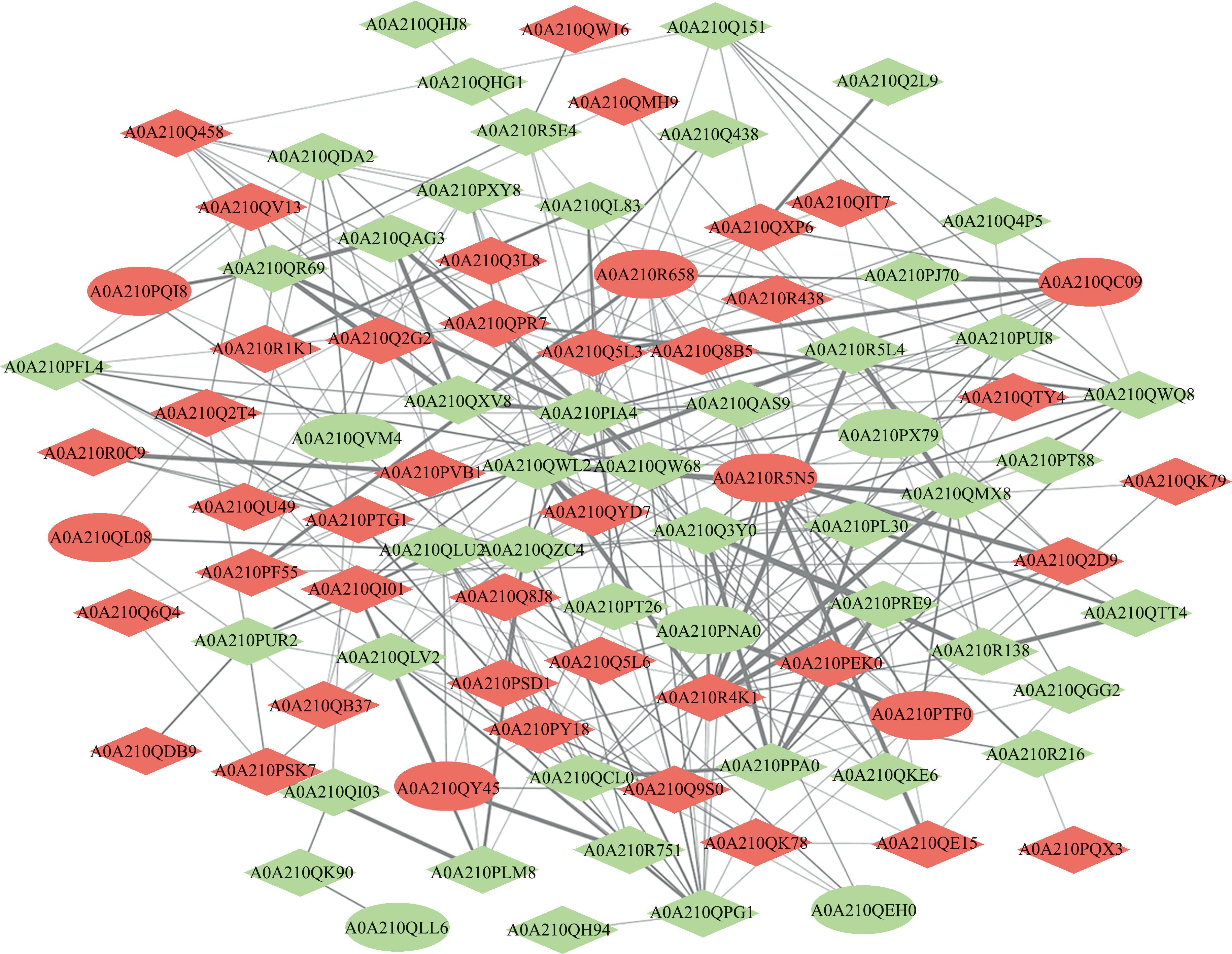
红色表示上调;绿色表示下调;椭圆形表示与壳色和矿化相关及与免疫、神经及甲基化调控相关的蛋白。
Red indicates up-regulation;and green indicates down-regulation;Ellipses represent proteins related to shell color and mineralization,as well as immune,neural,and methylation regulation.
图6 两种壳色虾夷扇贝差异表达蛋白互作网络
Fig.6 Protein-protein interaction (PPi) network for DEPs from Yesso scallops with two shell colour
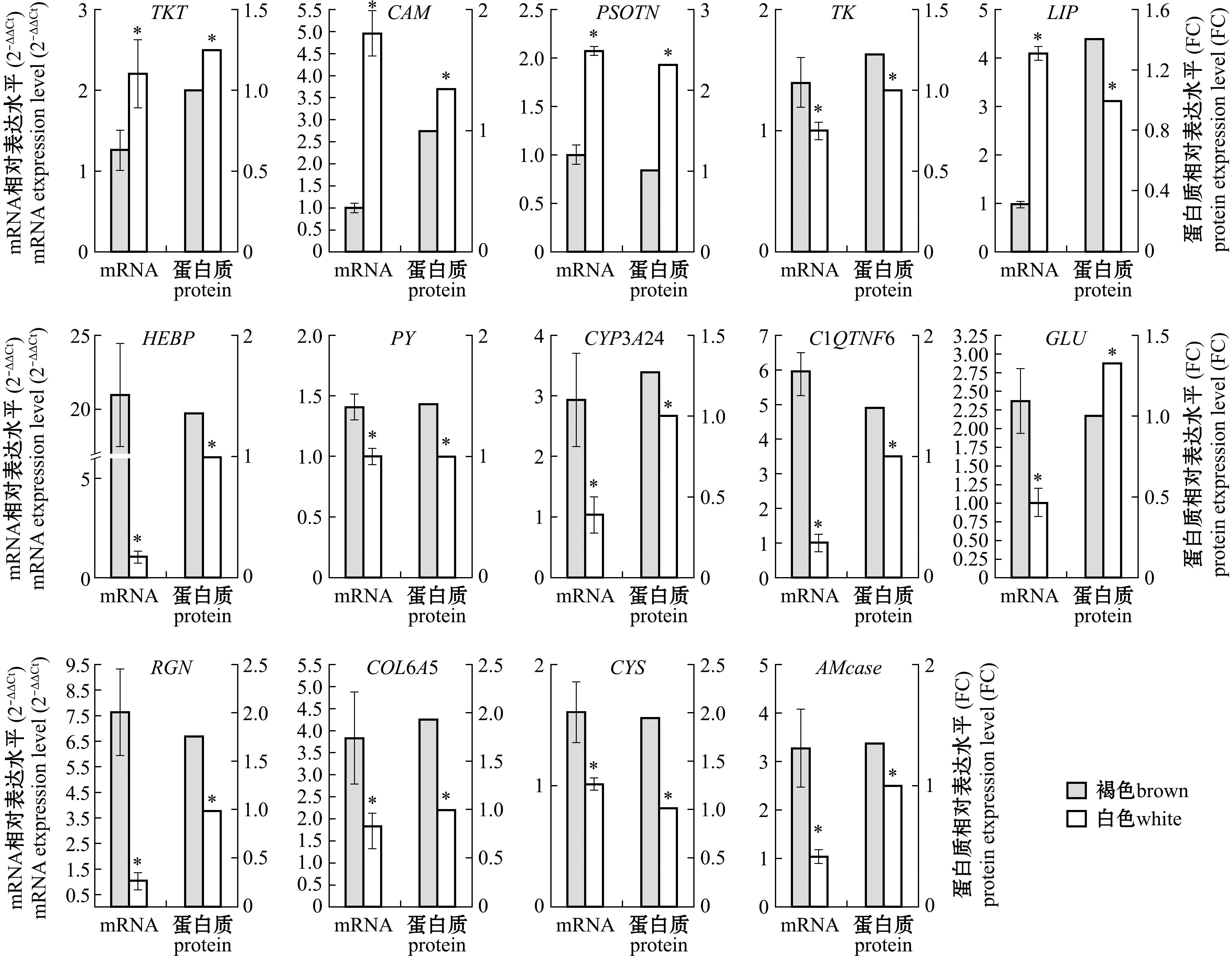
*表示差异显著(P value<0.05)。
*means significant differences (P value<0.05).
图7 差异表达蛋白在转录水平和蛋白水平的表达情况
Fig.7 Expressions of DEPs at transcriptional and protein levels
2.6 差异表达蛋白转录水平上的验证
利用qRT-PCR技术,对14个DEPs进行了差异表达蛋白转录水平的表达验证。从图5可见,12个DEPs与其对应的mRNA具有一致的表达变化趋势,且两种壳色扇贝间均存在显著性差异,2个DEPs的mRNA和蛋白质检测表现出相反的变化趋势,表明了蛋白质组学检测结果的可靠性。
3 讨论
蛋白质作为生物体中最重要的大分子有机化合物之一,是基因功能的关键执行者,对维持生命活动起着重要的作用。定量蛋白质组学,可以实现对样本中表达的全部蛋白质进行鉴定和定量,筛选样本或分组之间差异表达蛋白,并结合生物信息学分析预测差异蛋白功能,从而揭示蛋白质在细胞信号传导、代谢调控、生长发育等生命活动中的作用机制[22]。目前,贝壳壳色形成机制研究一直备受关注,但在蛋白质组层面开展的工作还相对有限。本研究中利用TMT标记定量蛋白质组学技术对两种不同壳色虾夷扇贝的外套膜组织进行蛋白质组学分析,挖掘了虾夷扇贝壳色形成的重要候选蛋白和分子通路,表明黑色素合成过程在虾夷扇贝壳色形成中发挥了重要作用,钙离子调控相关蛋白和通路很可能参与了虾夷扇贝黑色素合成的调控。本研究中从蛋白质组层面初步探索了虾夷扇贝壳色形成的分子基础和调控机制,为贝类壳色形成机制的深入解析提供了更多线索。
3.1 黑色素合成及其调控过程在壳色形成中的重要作用
贝类的贝壳主要由95%~99%的碳酸钙和不到5%的有机大分子组成。贝类的壳色一般由结合在壳质上的有机色素导致,如黑色素、类萝卜素、多烯类色素和四吡咯色素等[23]。其中,黑色素是自然界中最常见的生物色素之一。目前,已在虾夷扇贝等多种贝类贝壳中检测到黑色素的存在[24-25]。在多种贝类中,如菲律宾蛤仔(Ruditapes philippinarum)、长牡蛎(Crassostrea gigas)、黑蝶贝(Pinctada margaritifera)、马氏珍珠贝(Pinctada fucatamartensii)等,通过不同壳色个体外套膜的转录组表达谱分析发现,黑色素形成相关基因或分子通路显著差异表达,表明黑色素是参与贝类壳色形成的重要色素[26-29]。本研究中对两种壳色虾夷扇贝的差异表达蛋白的功能富集分析发现,黑色素形成直接相关的GO功能和KEGG通路显著富集,即黑色素生物合成过程(melanin biosynthetic process,GO:0042438)和黑色素生成通路(melanogenesis,ko04024),表明黑色素的合成在虾夷扇贝壳色形成中发挥重要作用。在脊椎动物中,黑色素的合成受多条信号通路的调控,比较常见的有cAMP依赖信号通路、Wnt信号通路和ERK信号通路[30]。本研究中,相关的调控功能和通路也在两种壳色DEPs中显著富集,如Wnt-蛋白结合(Wnt-protein binding,GO:0017147)、Wnt信号通路的负调控(negative regulation of Wnt signaling pathway,GO:0030178)、cAMP信号通路(cAMP signaling pathway;ko04024)等,其很可能参与了虾夷扇贝黑色素合成的调控进而影响壳色的形成。相似结果在菲律宾蛤仔、海湾扇贝、硬壳蛤等的转录组测序结果中也有报道[5,31-32]。
3.2 钙离子参与调控黑色素的合成对壳色形成的影响
钙离子的稳态平衡在真核细胞中至关重要。贝类贝壳主要由碳酸钙组成,钙离子代谢相关途径在壳及壳色的形成过程中发挥重要作用[33]。在不同壳色文蛤(Meretrix meretrix)的转录组测序中发现,钙离子信号通路可通过激活Notch信号通路控制壳色的产生[3]。在紫色和白色珍珠层三角帆蚌的转录组测序中鉴定获得多个钙离子代谢相关基因,如calreticulin,calmodulin,calcium-ATPase,calcium binding protein等,推测其可能调控三角帆蚌珍珠或珍珠层颜色的产生[12];在不同壳色马氏珍珠贝的比较转录组和蛋白质组学研究中,发现来自黑色素生成通路的多个calmodulin基因和蛋白显著差异表达,推测其影响了马氏珠母贝外套膜黑色素的合成[29]。Sun等[34]在虾夷扇贝外套膜的转录组数据中同样鉴定获得多个参与钙离子代谢的基因,如calmodulin、calmodulin-like protein等,推测其参与虾夷扇贝壳及壳色的形成。Buffey等[35]发现,钙离子和calmodulin信号系统能够抑制小鼠B16黑色素瘤细胞黑色素的合成,而钙离子对黑色素合成通路中酪氨酸酶活性同样具有抑制作用[36]。本研究中,钙离子信号通路(calcium signaling pathway,ko04020)在两种壳色虾夷扇贝DEPs中显著富集;此外,在DEPs中发现9个calmodulin和1个calnexin等钙离子调控相关蛋白在白色个体中显著上调。因此,推测钙离子调控过程很可能抑制了白色扇贝中黑色素的合成。
3.3 其他影响壳色形成的调控途径
扇贝外套膜中具有神经节和发达的神经纤维,调控扇贝的生命活动[37]。本研究中,与神经调控相关的GO功能和分子通路在两种壳色DEPs中也显示显著富集,表明壳色形成很可能是一个受神经调控的过程。此外,前期研究中发现,两种壳色虾夷扇贝外套膜全基因组DNA甲基化水平存在明显差异,黑色素合成和卟啉色素合成过程相关基因的表达受到DNA甲基化的调控[17]。本研究中检测到多个甲基转移酶在两种壳色中差异表达(表2),很可能参与调控了虾夷扇贝壳色形成相关基因的DNA甲基化水平,进而调节了壳色相关基因的表达水平并影响了壳色的形成。
3.4 贝类壳色与免疫间的关联
研究表明,贝类壳色与免疫力之间存在密切关系[2,38]。研究发现,黑壳马氏珠母贝呈现出更高的存活率和更强的免疫能力[38]。在对贝类不同壳色个体的组学研究中,同样发现与免疫有关的基因或过程显著差异表达,表明了贝类不同壳色个体间的免疫差异[5,34]。本研究中,免疫相关的GO功能和分子通路在两种壳色DEPs中显著富集,并鉴定获得多个与免疫相关的差异表达蛋白,分子层面表明了两种壳色虾夷扇贝间存在免疫差异,这为利用壳色进行抗性性状的定向改良提供了科学参考。
4 结论
1)本研究中采用TMT标记定量技术,对不同壳色虾夷扇贝外套膜进行蛋白质组学分析。经蛋白差异表达分析,共获得两种壳色扇贝间显著差异表达蛋白343个。
2)差异表达蛋白的GO和KEGG功能富集分析显示,黑色素合成、钙离子调控和神经调控相关功能和通路显著富集,推测其在虾夷扇贝壳色的形成及调控中发挥重要作用。免疫相关功能和分子通路也显示显著富集,表明两种壳色扇贝间很可能存在免疫差异。
3)qRT-PCR表明,DEPs的表达变化趋势与其相应的mRNA基本一致,证明了本试验中蛋白质组学检测结果的可靠性。
[1] WILLIAMS S T.Molluscan shell colour[J].Biological Reviews,2017,92(2):1039-1058.
[2] 丁鉴锋,杨霏,闫喜武,等.不同壳色菲律宾蛤仔免疫机能的比较研究[J].大连海洋大学学报,2012,27(5):411-416.DING J F,YANG F,YAN X W,et al.Comparison of the immune defense functions in Manila clam Ruditapes philippinarum with different shell colors[J].Journal of Dalian Ocean University,2012,27(5):411-416.(in Chinese)
[3] YUE X,NIE Q,XIAO G Q,et al.Transcriptome analysis of shell color-related genes in the clam Meretrix meretrix[J].Marine Biotechnology,2015,17(3):364-374.
[4] NIE H T,JIANG K Y,JIANG L W,et al.Transcriptome analysis reveals the pigmentation related genes in four different shell color strains of the Manila clam Ruditapes philippinarum[J].Genomics,2020,112(2):2011-2020.
[5] TENG W,CONG R H,QUE H Y,et al.De novo transcriptome sequencing reveals candidate genes involved in orange shell coloration of bay scallop Argopecten irradians[J].Journal of Oceanology and Limnology,2018,36(4):1408-1416.
[6] ASLAM B,BASIT M,NISAR M A,et al.Proteomics:technologies and their applications[J].Journal of Chromatographic Science,2017,55(2):182-196.
[7] SACO A,PANEBIANCO A,BLANCO S,et al.Integration of transcriptomics and proteomics improves the characterization of the role of mussel gills in a bacterial waterborne infection[J].Frontiers in Marine Science,2021,8:735309.
[8] SUN J,ZHANG Y,THIYAGARAJAN V,et al.Protein expression during the embryonic development of a gastropod[J].Proteomics,2010,10(14):2701-2711.
[9] DI G,LUO X,YOU W,et al.Proteomic analysis of muscle between hybrid abalone and parental lines Haliotis gigantea Reeve and Haliotis discus Hannai Ino[J].Heredity,2015,114(6):564-574.
[10] HUANG S Q,JIANG H J,ZHANG L,et al.Integrated proteomic and transcriptomic analysis reveals that polymorphic shell colors vary with melanin synthesis in Bellamya purificata snail[J].Journal of Proteomics,2021,230:103950.
[11] 王志新,梁海鹰,杜晓东,等.蛋白质组学在贝类研究中的应用[J].生命科学研究,2014,18(2):184-188.WANG Z X,LIANG H Y,DU X D,et al.Applications of proteomic techniques to molluscs research[J].Life Science Research,2014,18(2):184-188.(in Chinese)
[12] CHEN Y,LIU C,LI S G,et al.Repaired shells of the pearl oyster largely recapitulate normal prismatic layer growth:a proteomics study of shell matrix proteins[J].ACS Biomaterials Science &Engineering,2019,5(2):519-529.
[13] MARIE B,MARIE A,JACKSON D J,et al.Proteomic analysis of the organic matrix of the abalone Haliotis asinina calcified shell[J].Proteome Science,2010,8:54.
[14] KOSAKA Y.Scallops:Biology,Ecology,Aquaculture,and Fisheries[M].3rd.Amsterdam:Elsevier,2016:891-925.
[15] DING J,ZHAO L,CHANG Y Q,et al.Transcriptome sequencing and characterization of Japanese scallop Patinopecten yessoensis from different shell color lines[J].PLoS One,2015,10(2):e0116406.
[16] MAO J X,ZHANG W J,WANG X B,et al.Histological and expression differences among different mantle regions of the yesso scallop (Patinopecten yessoensis) provide insights into the molecular mechanisms of biomineralization and pigmentation[J].Marine Biotechnology,2019,21(5):683-696.
[17] YUAN C Z,MAO J X,SUN H Y,et al.Genome-wide DNA methylation profile changes associated with shell colouration in the Yesso scallop (Patinopecten yessoensis) as measured by whole-genome bisulfite sequencing[J].BMC Genomics,2021,22(1):740.
[18] WANG S,ZHANG J B,JIAO W Q,et al.Scallop genome provides insights into evolution of bilaterian karyotype and development[J].Nature Ecology &Evolution,2017,1(5):120.
[19] ASHBURNER M,BALL C A,BLAKE J A,et al.Gene ontology:tool for the unification of biology.The Gene Ontology Consortium[J].Nature Genetics,2000,25(1):25-29.
[20] KANEHISA M,GOTO S.KEGG:Kyoto encyclopedia of genes and genomes[J].Nucleic Acids Research,2000,28(1):27-30.
[21] 杭雲娜,王怡颖,孙红妍,等.虾夷扇贝 UROS 基因的全基因组鉴定,序列特征及表达分析[J].水产科学,2024,43(2):273-280.HANG Y N,WANG Y Y,SUN H Y,et al.Genome-Wide identificantion,characterization and expression of EROS gene in yesso scallop patinopecten yessoensis[J].Fisheries science,2024,43(2):273-280.(in Chinese)
[22] 薛淑群,刘伟,匡友谊.蛋白质组学研究的技术体系及进展[J].水产学杂志,2004,17(1):77-81.XUE S Q,LIU W,KUANG Y Y.Progress on proteome research[J].Chinese Journal of fisheries,2004,17(1):77-81.(in Chinese)
[23] WILLIAMS S T,LOCKYER A E,DYAL P,et al.Colorful seashells:identification of haem pathway genes associated with the synthesis of porphyrin shell color in marine snails[J].Ecology and Evolution,2017,7(23):10379-10397.
[24] 于文超,何成,武长路,等.长牡蛎(Crassostrea gigas)贝壳与外套膜中黑色素的提取和鉴定[J].海洋与湖沼,2015,46(4):909-914.YU W C,HE C,WU C L,et al.Extraction and identification of melanin in shell and mantle of Pacific oyster Crassostrea gigas[J].Oceanologia et Limnologia Sinica,2015,46(4):909-914.(in Chinese)
[25] SUN X J,WU B,ZHOU L Q,et al.Isolation and characterization of melanin pigment from yesso scallop Patinopecten yessoensis[J].Journal of Ocean University of China,2017,16(2):279-284.
[26] FENG D D,LI Q,YU H,et al.Comparative transcriptome analysis of the Pacific oyster Crassostrea gigas characterized by shell colors:identification of genetic bases potentially involved in pigmentation[J].PLoS One,2015,10(12):e0145257.
[27] LEMER S,SAULNIER D,GUEGUEN Y,et al.Identification of genes associated with shell color in the black-lipped pearl oyster,Pinctada margaritifera[J].BMC Genomics,2015,16(1):568.
[28] XU M,HUANG J,SHI Y,et al.Comparative transcriptomic and proteomic analysis of yellow shell and black shell pearl oysters,Pinctada fucata martensii[J].BMC Genomics,2019,20(1):469.
[29] DING J F,WEN Q,HUO Z M,et al.Identification of shell-color-related microRNAs in the Manila clam Ruditapes philippinarum using high-throughput sequencing of small RNA transcriptomes[J].Scientific Reports,2021,11(1):8044.
[30] CHANG T S.Natural melanogenesis inhibitors acting through the down-regulation of tyrosinase activity[J].Materials,2012,5(9):1661-1685.
[31] BAI Y T,NIE H T,WANG Z X,et al.Genome-wide identification and transcriptome-based expression profiling of Wnt gene family in Ruditapes philippinarum[J].Comparative Biochemistry and Physiology Part D:Genomics and Proteomics,2020,35:100709.
[32] HU Z,SONG H,YANG M J,et al.Transcriptome analysis of shell color-related genes in the hard clam Mercenaria mercenaria[J].Comparative Biochemistry and Physiology Part D:Genomics and Proteomics,2019,31:100598.
[33] ANNE S V.The molluscan shell secretome:unlocking calcium pathways in a changing world[D].Edinburgh,Scotland,UK:Heriot-Watt University,2017
[34] SUN X J,YANG A G,WU B,et al.Characterization of the mantle transcriptome of yesso scallop (Patinopecten yessoensis):identification of genes potentially involved in biomineralization and pigmentation[J].PLoS One,2015,10(4):e0122967.
[35] BUFFEY J,THODY A J,BLEEHEN S S,et al.Alpha-melanocyte-stimulating hormone stimulates protein kinase C activity in murine B16 melanoma[J].The Journal of Endocrinology,1992,133(3):333-340.
[36] S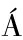 NCHEZ-FERRER
NCHEZ-FERRER 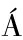 ,NEPTUNO RODR
,NEPTUNO RODR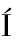 GUEZ-L
GUEZ-L PEZ J,GARC
PEZ J,GARC A-C
A-C NOVAS F,et al.Tyrosinase:a comprehensive review of its mechanism[J].Biochimica et Biophysica Acta (BBA)-Protein Structure and Molecular Enzymology,1995,1247(1):1-11.
NOVAS F,et al.Tyrosinase:a comprehensive review of its mechanism[J].Biochimica et Biophysica Acta (BBA)-Protein Structure and Molecular Enzymology,1995,1247(1):1-11.
[37] ZHANG W J,MAO J X,YUAN C Z,et al.Histological changes in the mantle tissue of the yesso scallop Patinopecten yessoensis shell infested by Polydora[J].Journal of Shellfish Research,2020,39(1):87.
[38] ADZIGBLI L,WANG Z M,LI J H,et al.Survival,retention rate and immunity of the black shell colored stocks of pearl oyster Pinctada fucata martensii after grafting operation[J].Fish &Shellfish Immunology,2020,98:691-698.