肌源性调节因子(myogenic regulatory factors,MRF)可以在胚胎发育过程中调节肌肉纤维数量[1]。MRF家族包括4个成员,分别为肌源性调节因子D(MyoD)、G(MyoG)、5(MYF5)和6(MYF6)。所有MRF都参与肌肉发育,并促进成肌细胞的增殖与分化及成熟后的功能[2]。在MRF家族基因中,MYF5最早在胚胎肌肉发育过程中表达[3],该基因也在其他组织中表达,包括前脂肪细胞和神经元[4],尽管其在这些组织中的表达低于肌肉组织[5]。骨骼肌干细胞中MyoD和MYF5基因缺失会导致肌源性编程改变和再生失败[6]。Zhao等[7]在秦川牛MYF5基因中发现,对秦川肉牛生长性能和品质性状有显著影响的单核苷酸多态性(SNP)位点有3个(g.5785C>T、g.5816A>G和g.6535A>G),H1H3二倍体体型较大,牛肉品质较好。MYF5基因变异与中国草地短尾羊(GSTS)生长性能和屠宰性状有显著相关关系:7个SNP位点中,有5个位点对生长性能和屠宰性状有显著影响;H1H3和H2H3二倍体具有更高的体质量和更大的体型;单倍型H3在肉类生产方面的表现优于其他方面[8]。Tang等[9]发现,京海黄鸡MYF5基因的8个SNP位点和单倍型均与生长、繁殖性状相关。Wang等[10]发现,MYF5基因多态性对伊拉和天府黑兔肉质性状有显著性影响。在鱼类MYF5基因方面的研究,仅郭玉函等[11-12]克隆了大口黑鲈(Micropterus salmoides)MYF5基因,并进行了序列和功能分析;钟茂春等[13]用单链构象多态性(SSCP)方法分析了MYF5基因在珠江水系3个不同江段鲮(Cirrhinus molitorella)中的遗传变异,结果表明,鲮MYF5基因的C412T位点与个体生长有相关性,可作为候选基因标记,用于鲮生长相关分子标记辅助育种研究。MYF5基因与其他鱼类生长性状的关联分析尚未见报道。
罗非鱼是世界上最重要的养殖鱼类之一,中国是罗非鱼养殖产量最大的国家,占世界罗非鱼养殖产量的近一半。2020年和2021年,中国罗非鱼养殖产量分别为166.5万t和166.2万t,在国民经济中占有重要地位(据《2022年渔业统计年鉴》)。尽管罗非鱼的年产量相当可观,但在相同的养殖环境中,个体的生长表现不同,个体间的体质量和形态特征存在显著性差异。这种情况影响了产品的规格和质量[14],因此,开展罗非鱼生长相关位点筛选,辅助罗非鱼生长性状选育研究具有重要意义。鉴于MYF5基因多态性与家畜的生长和肉质性状相关,本研究中,通过PCR及测序法筛查尼罗罗非鱼(Oerochromis niloticus)MYF5基因5′UTR、外显子、内含子和3′UTR区的SNP位点,并与生长数据相结合,在尼罗罗非鱼高要亲代群体中筛选与生长性状相关的SNP位点,并将这些SNP位点在高要子代群体、番禺群体及海南群体中验证其普适性,以期为罗非鱼养殖中以生长性状为目的的选育提供有效的分子标记。
1 材料与方法
1.1 材料
用于筛查 SNP位点的样本来源于珠江水产研究所水产良种基地养殖的新吉富尼罗罗非鱼(高要群体),取40尾鱼的鳍条样本,在20 ℃下保存于无水乙醇中。
用于生长性状关联分析的亲代群体及子代群体样品分别为珠江水产研究所水产良种基地的新吉富尼罗罗非鱼(高要群体)、广东罗非鱼良种场的广特超罗非鱼(番禺群体)及海南省水产科学院的尼罗罗非鱼(海南群体)。高要亲代群体为2020年10月繁殖,在塘养殖越冬至次年7月;高要子代群体为2021年的4月繁殖,取200尾鱼苗在同一水泥池养殖3个月(经激素处理过,全雄);番禺群体为2021年4月繁殖(经激素处理过,全雄),取200尾鱼苗在同一水泥池养殖3个月;海南群体为2021年的春苗1万尾,放至33.4 hm2的土塘中养殖3个月(未进行激素处理正常雌雄群体)。测量高要子代群体162尾,测量番禺群体175尾,测量海南群体300尾(雌、雄各150尾),测量体质量、全长、体长、头长、体高和体宽。
1.2 方法
1.2.1 基因组DNA的提取 采用磁珠法基因组DNA提取试剂盒(NanoMagBio)提取基因组DNA,具体方法步骤参照试剂盒说明书。用琼脂糖凝胶电泳检测基因组DNA提取质量。
1.2.2 MYF5基因SNP位点的筛查 根据MYF5基因[GenBank登录号为NC_031981.2(15815227~15818324,complement)]区域序列,利用Primer Primer 5.0软件设计3对上、下游引物:MYF5-1-F(TTAGATCTGGCCAATGTGGG),MYF5-1-R(GTGCAGAACATGCAGTCAGG);MYF5-2-F(GAATCTCCGTCCTCTGCTTG),MYF5-2-R(TAAATATGTCTCACTGGGGATAATAG);MYF5-3-F(TCCTGTGAAGGAAACCTTGC),MYF5-3-R(TCACGTTTGGTAGGAAGGAAC),MYF5-3-SEQ(CACAGAGTGCAGCTTTAC)。以高要亲代群体中40个个体的DNA为模板进行PCR扩增。PCR反应体系(共30 μL):2×Taq PCR Master Mix 15 μL,基因组DNA(20 ng)1 μL,上、下游引物(10 pmol/μL)各1 μL,ddH2O 12 μL。反应条件:95 ℃下预变性5 min;95 ℃下变性30 s,60 ℃下退火30 s,72 ℃下延伸45 s,共进行35个循环;最后在72 ℃下再延伸5 min,16 ℃下退火1 min,PCR产物于4 ℃下保存。PCR产物送Sanger公司测序,测序平台为ABI公司的3730xl DNA Analyzer测序仪。将测序良好的全长序列在SeqMan软件中进行多序列比对,筛查SNP位点。
1.2.3 MYF5基因SNP位点的分型 从筛查到的35个SNP位点中筛选多态性较好的30个位点进行分型分析。SNP位点分型采用“1.2.2节”中PCR产物测序法,反应体系和反应条件同“1.2.2节”。首先对高要亲代群体(213尾)进行分型,再将获得的与生长性状相关的SNP位点在高要子代群体(162尾)及番禺群体(175尾)中进行分型,以验证亲子代群体间生长相关SNP位点的可遗传性,以及不同群体间的可重复性。
1.2.4 MYF5基因SNP位点在海南群体的分型验证 将与高要亲代群体、高要子代群体及番禺群体生长性状相关的位点,在海南群体(雌、雄各150尾)中进行分型分析,并与生长性状进行关联分析,进一步验证所获得SNP位点的可重复性。
1.2.5 遗传信息统计 利用PIC-CALC 0.6对SNP位点的多态信息含量(polymorphism information content,PIC)进行分析,PIC<0.25时SNP属于低度多态性,0.25≤PIC≤0.50时SNP属于中度多态性,PIC>0.50时SNP属于高度多态性。利用Popgene 3.2软件进行哈迪-温伯格平衡(Hardy-Weinberg equilibrium,HWE)状态的卡方检验,并计算观测杂合度(observed heterozygosity,Ho)、期望杂合度(expected heterozygosity,He)、有效等位基因数(effective number of alleles,Ne)、基因频率及等位基因频率。采用HaploView 4.2软件对SNP位点进行连锁不平衡分析和单倍块(haplotype block)构建。利用单倍型手动分析出双倍型。
1.3 数据处理
生长性状测量数据用平均值±标准差表示。采用 SPSS 19软件一般线性模型(general linear model,GLM)中的多元方差分析模块进行SNP位点不同基因型及双倍型与6 个生长性状的关联分析。
2 结果分析
2.1 MYF5基因部分序列SNP位点的获得
将PCR扩增获得的3 098 bp的片段(GenBank accession No.NC_031987(15815227~15818324,complement)进行序列比对,获得35个SNP位点。从该片段的第一个碱基算起,分别命名为S1、S2、S3、…、S35(图1)。

图1 MYF5基因SNP位点的分布位置
Fig.1 SNP locus distribution of MYF5 gene
2.2 MYF5基因SNP位点在各群体中的遗传多样性
2.2.1 MYF5基因SNP位点在高要亲代群体中的遗传多样性 MYF5基因 30个SNP位点在高要亲代群体中的多样性分析显示:高要亲代群体平均Ne为1.446 6,平均Ho为0.263 3,平均He为0.268 6,平均PIC为0.214 9;高要亲代群体的Ne分布为1.023 9~2.000 0,Ho分布为0.023 6~0.495 3,He分布为0.023 4~0.501 2;高要亲代群体的PIC分布为0.023 0~0.375 0,其中,15个位点属于低度多态性,15个位点属于中度多态性;S18位点在群体中偏离Hardy-Weinberg平衡定律,其余位点均符合Hardy-Weinberg平衡定律(表1)。
表1 MYF5基因30个SNP位点在尼罗罗非鱼高要亲代群体的遗传信息
Tab.1 Genetic information of 30 SNP loci of MYF5 gene in Gaoyao parent population of Oreochromis niloticus

注:*表示偏离Hardy-Weinberg定律;*,P<0.05,**,P<0.01,***,P<0.001;基因型频率单位为%,下同。
Note:*,deviation from Hardy-Weinberg equilibrium;*,P<0.05,**,P<0.01,***,P<0.001;genotype frequency unit is %,et sequentia.
SNP位点(变异位置)locus(variation site)有效等位基因数Ne期望杂合度He观测杂合度Ho多态信息含量PIC哈温平衡H-W基因型(样本数/基因型频率)genotype (sample number/genotype frequency)S1(C-26T)1.997 20.500 50.490 60.375 00.751CC(58/27)CT(104/49)TT(50/24)S2(C-65T)1.766 90.435 10.419 80.339 00.611CC(58/27)CT(104/49)TT(50/24)S3(A-113G)1.594 00.373 50.372 60.302 00.982AA(79/37)AG(13/6)GG(120/57)S4(C-170T)1.594 00.373 50.372 60.302 00.982CC(121/57 )CT(79/37)TT(13/6)S5(C-323T)1.274 20.215 70.217 00.191 00.912CC(163/77)CT(46/22)TT(3/1)S6(C-563G)1.766 90.435 10.419 80.339 00.611CC(23/11)CG(89/42)GG(100/47)S7(A-626T)1.865 20.465 00.495 30.356 00.344AA(82/39)AT(105/50)TT(25/12)S8(A-645G)1.653 80.396 30.382 10.316 00.607AA(17/8)AG(81/38)GG(114/54)S9(C-738T)1.189 50.159 70.165 10.146 00.600CC(1/0.4)CT(35/17)TT(176/83)S10(C-754G)1.068 20.064 00.066 00.062 00.620CC(198/93)CG(14/7)GG(0/0)S11(A-794G)1.557 50.358 80.353 80.293 00.849AA(125/59)AG(75/35)GG(12/6)S13(A-984T)1.043 40.041 60.042 50.041 00.753AA(0/0)AG(25/12)GG(187/88)S14(A-1241G)1.124 80.111 20.117 90.104 00.363AA(167/78)AT(24/11)TT(22/10)S15(C-1367T)1.119 60.107 10.113 20.101 00.384CC(188/89)CT(24/11)TT(0/0)S16(C-1376T)1.023 90.023 40.023 60.023 00.862CC(0/0)TC(5/2)TT(207/98)S17(A-1567C)1.033 60.032 60.033 00.032 00.807AA(2/1)AC(7/3)CC(205/96)S18(C-1739G)1.189 50.159 70.061 30.146 00.000∗∗∗CC(12/6)CG(13/6)GG(187/88)S19(C-1774T)1.234 20.190 20.202 80.169 00.340CC(168/80)CT(43/20)TT(1/0.4)S20(A-2072G)1.033 60.032 60.033 00.032 00.807AA(2/1)AG(7/3)GG(205/96)S21(C-2090G)1.968 10.493 10.485 80.371 00.764CC(41/19)CG(103/49)GG(68/32)S22(A-2117G)2.000 00.501 20.462 30.375 00.336AA(57/27)AG(98/46)GG(57/27)S23(A-2221G)1.581 80.368 70.391 50.294 00.644AA(10/5)AG(83/39)GG(119/56)S24(C-2345T)1.985 70.497 60.434 00.375 00.899CC(69/33)CT(92/43)TT(51/24)S25(C-2349T)1.453 50.312 70.292 50.252 00.117AA(10/5)AT(62/29)TT(140/66)S26(C-2445G)1.326 90.246 90.268 90.200 00.376CC(153/72)CG(57/27)GG(2/1)S30(A-2659G)1.028 70.028 0 0.028 30.028 00.834AA(0/0)AG(6/3)GG(206/97)S31(A-2746T)1.088 50.081 50.056 60.062 00.099AA(197/93)AT(12/6)TT(3/1)S32(G-2759T)1.974 70.494 80.490 60.373 00.950GG(42/20)GT(104/49)TT(66/31)S34(C-2884T)1.677 20.404 70.448 10.324 00.099CC(105/50)CT(95/45)TT(12/6)S35(A-2938G)1.184 00.155 80.160 40.123 00.066AA(1/0.4)AG(34/16)GG(177/83)平均 average1.446 60.268 60.263 30.214 9
2.2.2 MYF5基因SNP位点在高要子代群体中的遗传多样性 MYF5基因14个SNP位点在高要子代群体中的多样性分析显示:高要子代群体平均Ne为1.494 0,平均Ho为0.267 6,平均He为0.300 4,平均PIC为0.374 5;高要子代群体的Ne分布为1.000 0~1.975 6,Ho分布为0~0.456 8,He分布为0~0.495 4;高要子代群体的PIC分布为0~0.571 3,其中,2个位点属于低度多态性,8个位点属于中度多态性,4个位点属于高度多态性;9个位点在群体中均符合Hardy-Weinberg平衡定律,其余位点偏离Hardy-Weinberg平衡定律(表2)。
表2 MYF5基因14个SNP位点在尼罗罗非鱼高要子代群体的遗传信息
Tab.2 Genetic information of 14 SNP loci of MYF5 gene in Gaoyao offspring population of Oreochromis niloticus
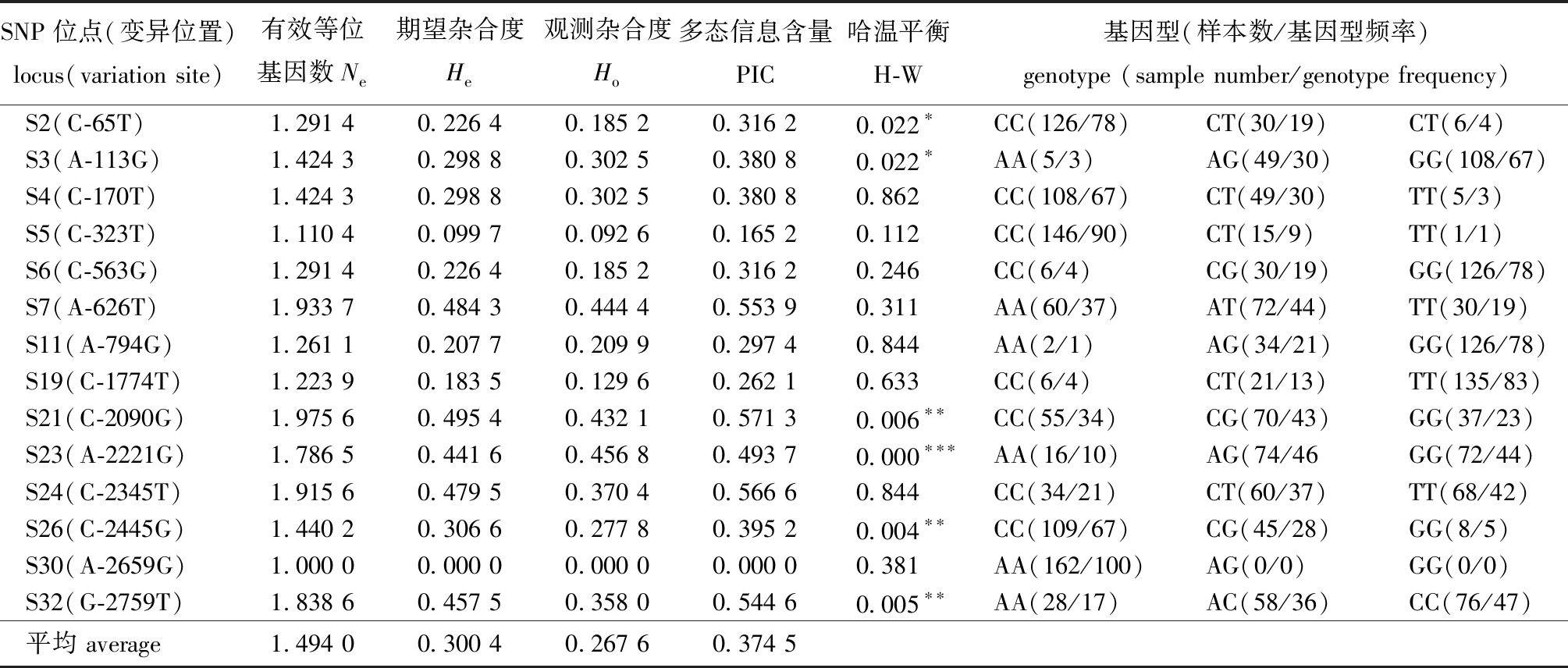
SNP位点(变异位置)locus(variation site)有效等位基因数Ne期望杂合度He观测杂合度Ho多态信息含量PIC哈温平衡H-W基因型(样本数/基因型频率)genotype (sample number/genotype frequency)S2(C-65T)1.291 40.226 40.185 20.316 20.022∗CC(126/78)CT(30/19)CT(6/4)S3(A-113G)1.424 30.298 80.302 50.380 80.022∗AA(5/3)AG(49/30)GG(108/67)S4(C-170T)1.424 30.298 80.302 50.380 80.862CC(108/67)CT(49/30)TT(5/3)S5(C-323T)1.110 40.099 70.092 60.165 20.112CC(146/90)CT(15/9)TT(1/1)S6(C-563G)1.291 40.226 40.185 20.316 20.246CC(6/4)CG(30/19)GG(126/78)S7(A-626T)1.933 70.484 30.444 40.553 90.311AA(60/37)AT(72/44)TT(30/19)S11(A-794G)1.261 10.207 70.209 90.297 40.844AA(2/1)AG(34/21)GG(126/78)S19(C-1774T)1.223 90.183 50.129 60.262 10.633CC(6/4)CT(21/13)TT(135/83)S21(C-2090G)1.975 60.495 40.432 10.571 30.006∗∗CC(55/34)CG(70/43)GG(37/23)S23(A-2221G)1.786 50.441 60.456 80.493 70.000∗∗∗AA(16/10)AG(74/46GG(72/44)S24(C-2345T)1.915 60.479 50.370 40.566 60.844CC(34/21)CT(60/37)TT(68/42)S26(C-2445G)1.440 20.306 60.277 80.395 20.004∗∗CC(109/67)CG(45/28)GG(8/5)S30(A-2659G)1.000 00.000 00.000 00.000 00.381AA(162/100)AG(0/0)GG(0/0)S32(G-2759T)1.838 60.457 50.358 00.544 60.005∗∗AA(28/17)AC(58/36)CC(76/47)平均 average1.494 00.300 40.267 60.374 5
2.2.3 MYF5基因SNP位点在番禺群体中的遗传多样性 MYF5基因 14 个 SNP 位点在番禺群体中的多样性分析显示:番禺群体平均Ne为1.548 2,平均Ho为0.324 5,平均He为0.330 8,平均PIC为0.396 6;番禺群体的Ne分布为1.017 3~1.985 4,He分布为0.017 0~0.497 7,Ho分布为0.017 1~0.542 9;番禺群体的PIC分布为0.033 1~0.566 6,其中,1个位点属于低度多态性,11个位点属于中度多态性,2个位点属于高度多态性;11个位点在群体中均符合Hardy-Weinberg平衡定律,3个位点偏离Hardy-Weinberg平衡定律(表3)。
表3 MYF5基因14个SNP位点在尼罗罗非鱼番禺群体的遗传信息
Tab.3 Genetic information of 14 SNP loci of MYF5 gene in Panyu population of Oreochromis niloticus
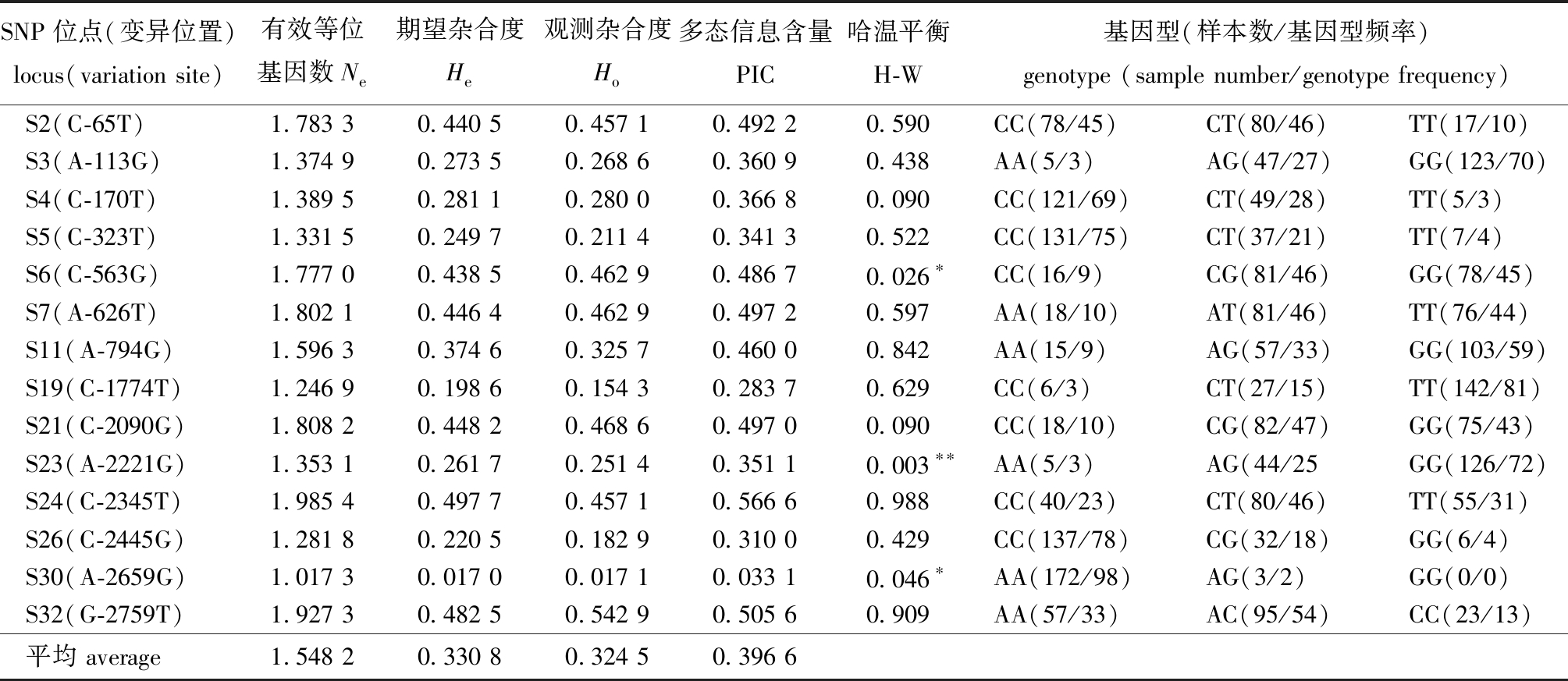
SNP位点(变异位置)locus(variation site)有效等位基因数Ne期望杂合度He观测杂合度Ho多态信息含量PIC哈温平衡H-W基因型(样本数/基因型频率)genotype (sample number/genotype frequency)S2(C-65T)1.783 30.440 50.457 10.492 20.590CC(78/45)CT(80/46)TT(17/10)S3(A-113G)1.374 90.273 50.268 60.360 90.438AA(5/3)AG(47/27)GG(123/70)S4(C-170T)1.389 50.281 10.280 00.366 80.090CC(121/69)CT(49/28)TT(5/3)S5(C-323T)1.331 50.249 70.211 40.341 30.522CC(131/75)CT(37/21)TT(7/4)S6(C-563G)1.777 00.438 50.462 90.486 70.026∗CC(16/9)CG(81/46)GG(78/45)S7(A-626T)1.802 10.446 40.462 90.497 20.597AA(18/10)AT(81/46)TT(76/44)S11(A-794G)1.596 30.374 60.325 70.460 00.842AA(15/9)AG(57/33)GG(103/59)S19(C-1774T)1.246 90.198 60.154 30.283 70.629CC(6/3)CT(27/15)TT(142/81)S21(C-2090G)1.808 20.448 20.468 60.497 00.090CC(18/10)CG(82/47)GG(75/43)S23(A-2221G)1.353 10.261 70.251 40.351 10.003∗∗AA(5/3)AG(44/25GG(126/72)S24(C-2345T)1.985 40.497 70.457 10.566 60.988CC(40/23)CT(80/46)TT(55/31)S26(C-2445G)1.281 80.220 50.182 90.310 00.429CC(137/78)CG(32/18)GG(6/4)S30(A-2659G)1.017 30.017 00.017 10.033 10.046∗AA(172/98)AG(3/2)GG(0/0)S32(G-2759T)1.927 30.482 50.542 90.505 60.909AA(57/33)AC(95/54)CC(23/13)平均 average1.548 20.330 80.324 50.396 6
2.2.4 MYF5基因SNP位点在海南群体中的遗传多样性 MYF5基因9个 SNP位点在海南群体雌性个体中的多样性分析显示:平均Ne为1.481 8,平均Ho为0.259 3,平均He为0.289 2,平均PIC为0.233 0;9个位点中,有4个位点属于低度多态性,5个位点属于中度多态性;8个位点在群体中均符合Hardy-Weinberg平衡定律,1个位点偏离Hardy-Weinberg平衡定律(表4)。
表4 MYF5基因9个SNP位点在尼罗罗非鱼海南雌性群体的遗传信息
Tab.4 Genetic information of 9 SNP loci of MYF5 gene in Hainan female population of Oreochromis niloticus
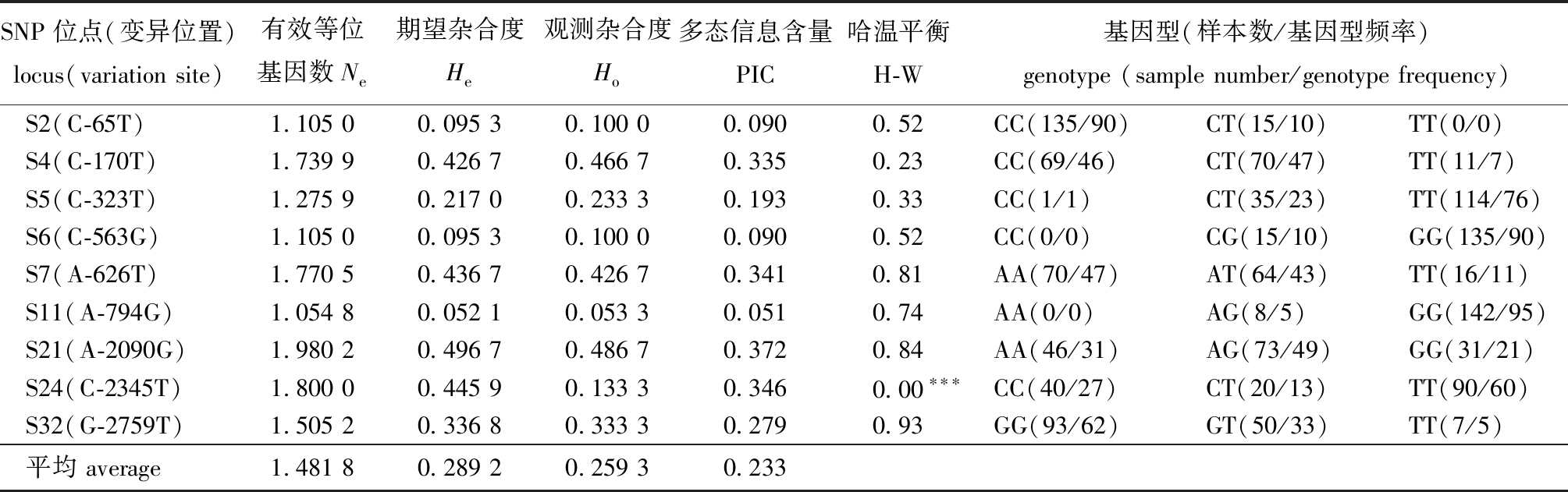
SNP位点(变异位置)locus(variation site)有效等位基因数Ne期望杂合度He观测杂合度Ho多态信息含量PIC哈温平衡H-W基因型(样本数/基因型频率)genotype (sample number/genotype frequency)S2(C-65T)1.105 00.095 30.100 00.0900.52CC(135/90)CT(15/10)TT(0/0)S4(C-170T)1.739 90.426 70.466 70.3350.23CC(69/46)CT(70/47)TT(11/7)S5(C-323T)1.275 90.217 00.233 30.1930.33CC(1/1)CT(35/23)TT(114/76)S6(C-563G)1.105 00.095 30.100 00.0900.52CC(0/0)CG(15/10)GG(135/90)S7(A-626T)1.770 50.436 70.426 70.3410.81AA(70/47)AT(64/43)TT(16/11)S11(A-794G)1.054 80.052 10.053 30.0510.74AA(0/0)AG(8/5)GG(142/95)S21(A-2090G)1.980 20.496 70.486 70.3720.84AA(46/31)AG(73/49)GG(31/21)S24(C-2345T)1.800 00.445 90.133 30.3460.00∗∗∗CC(40/27)CT(20/13)TT(90/60)S32(G-2759T)1.505 20.336 80.333 30.2790.93GG(93/62)GT(50/33)TT(7/5)平均 average1.481 80.289 20.259 30.233
MYF5基因9个SNP位点在海南群体雄性个体中的多样性比较丰富,平均Ne为1.478 1,平均Ho为0.274 8,平均He为0.290 9,平均PIC为0.236 0;9个位点中,4个位点属于低度多态性,5个位点属于中度多态性;8个位点在群体中均符合Hardy-Weinberg平衡定律,1个位点偏离Hardy-Weinberg平衡定律(表5)。
表5 MYF5基因9个SNP位点在尼罗罗非鱼海南雄性群体的遗传信息
Tab.5 Genetic information of 9 SNP loci of MYF5 gene in Hainan male population of Oreochromis niloticus

SNP位点(变异位置)locus(variation site)有效等位基因数Ne期望杂合度He观测杂合度Ho多态信息含量 PIC哈温平衡H-W基因型(样本数/基因型频率)genotype (sample number/genotype frequency)S2(C-65T)1.097 70.089 30.080 00.0850.22CC(137/91)CT(12/8)TT(1/1)S4(C-170T)1.574 30.366 00.386 70.2980.46CC(85/57)CT(58/39)TT(7/5)S5(C-323T)1.284 10.222 00.226 70.1970.76CC(2/1)CT(34/23)TT(114/76)S6(C-563G)1.090 40.083 20.073 30.0800.16CC(1/1)CG(11/7)GG(138/92)S7(A-626T)1.792 80.443 70.473 30.3440.39AA(65/43)AT(71/47)TT(14/9)S11(A-794G)1.134 60.119 00.113 30.1120.58AA(1/1)AG(17/11)GG(132/88)S21(A-2090G)1.997 80.501 10.526 70.3750.50AA(38/25)AG(79/53)GG(33/22)S24(C-2345T)1.800 00.445 90.280 00.3460.00∗∗∗CC(29/19)CT(42/28)TT(79/53)S32(G-2759T)1.531 20.348 10.313 30.2870.24GG(93/62)GT(47/31)TT(10/7)平均 average1.478 10.290 90.274 80.236
2.3 MYF5基因SNP位点与各群体生长性状的相关性
2.3.1 MYF5基因SNP位点与高要亲代群体生长性状的关联分析 从表6可见:S3位点AG、GG基因型个体的全长显著大于AA基因型个体(P<0.05);S4位点CC、CT基因型个体的全长显著大于TT基因型个体(P<0.05);S5位点CT基因型个体的头长显著大于TT基因型个体(P<0.05);S18位点CC基因型个体的体质量和体宽显著大于CG基因型个体(P<0.05);S24位点CT基因型个体的头长显著大于TT基因型个体(P<0.05)。
表6 MYF5基因中各SNP位点与高要亲代群体生长性状的关联分析
Tab.6 Association analysis of SNP locus in MYF5 gene with growth traits of Gaoyao parent population
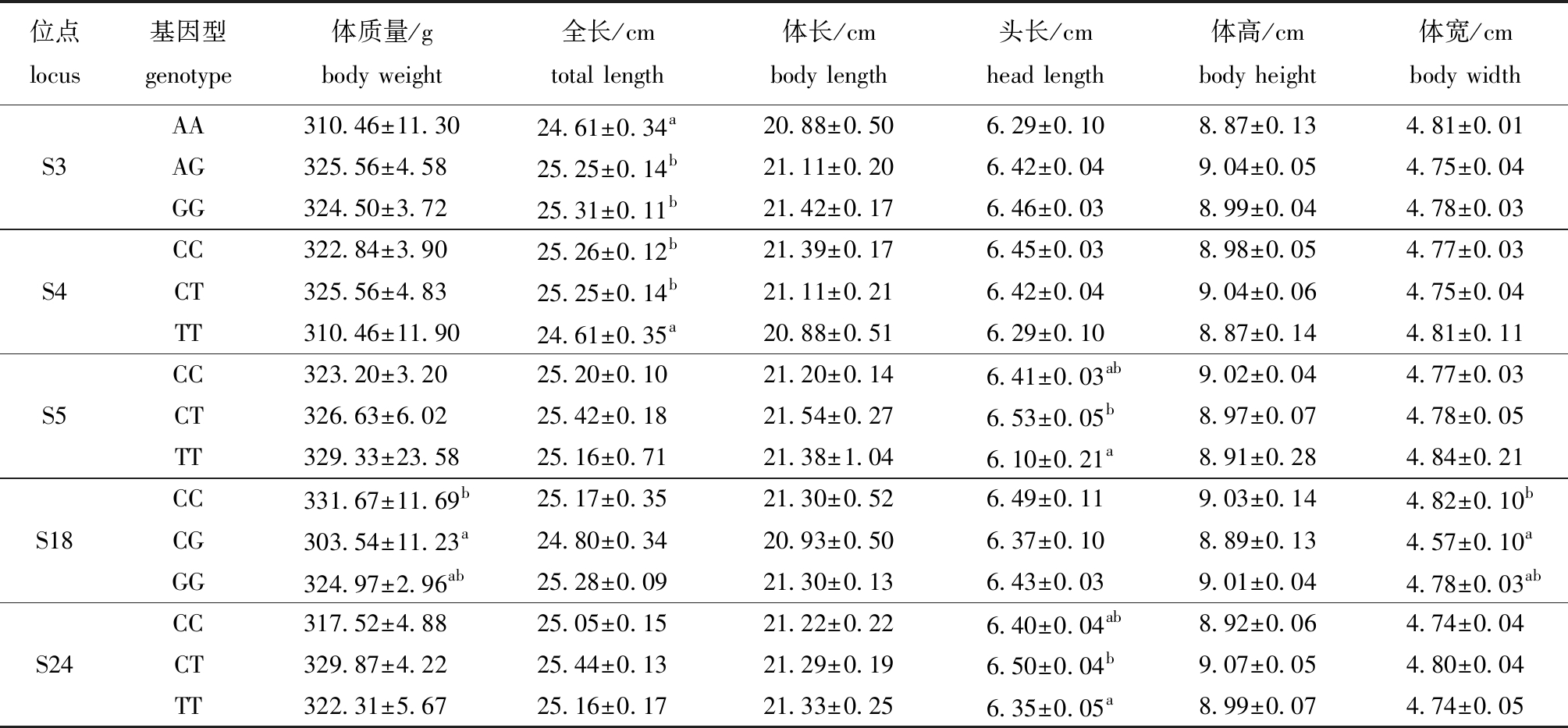
位点locus基因型genotype体质量/gbody weight全长/cmtotal length体长/cmbody length头长/cmhead length体高/cmbody height体宽/cmbody widthAA310.46±11.3024.61±0.34a20.88±0.506.29±0.108.87±0.134.81±0.01S3AG325.56±4.5825.25±0.14b21.11±0.206.42±0.049.04±0.054.75±0.04GG324.50±3.7225.31±0.11b21.42±0.176.46±0.038.99±0.044.78±0.03CC322.84±3.9025.26±0.12b21.39±0.176.45±0.038.98±0.054.77±0.03S4CT325.56±4.8325.25±0.14b21.11±0.216.42±0.049.04±0.064.75±0.04TT310.46±11.9024.61±0.35a20.88±0.516.29±0.108.87±0.144.81±0.11CC323.20±3.2025.20±0.1021.20±0.146.41±0.03ab9.02±0.044.77±0.03S5CT326.63±6.0225.42±0.1821.54±0.276.53±0.05b8.97±0.074.78±0.05TT329.33±23.5825.16±0.7121.38±1.046.10±0.21a8.91±0.284.84±0.21CC331.67±11.69b25.17±0.3521.30±0.526.49±0.119.03±0.144.82±0.10bS18CG303.54±11.23a24.80±0.3420.93±0.506.37±0.108.89±0.134.57±0.10aGG324.97±2.96ab25.28±0.0921.30±0.136.43±0.039.01±0.044.78±0.03abCC317.52±4.8825.05±0.1521.22±0.226.40±0.04ab8.92±0.064.74±0.04S24CT329.87±4.2225.44±0.1321.29±0.196.50±0.04b9.07±0.054.80±0.04TT322.31±5.6725.16±0.1721.33±0.256.35±0.05a8.99±0.074.74±0.05
2.3.2 MYF5基因SNP位点与高要子代群体生长性状的关联分析 从表7可见:S3位点AG基因型个体的体质量显著大于AA基因型个体(P<0.05);S4位点CT基因型个体的体质量显著大于TT基因型个体(P<0.05);S24位点CT基因型个体的体长、体高显著大于TT基因型个体(P<0.05)。
表7 MYF5基因中各SNP位点与高要子代群体生长性状的关联分析
Tab.7 Association analysis of SNP locus in MYF5 gene with growth traits of Gaoyao offspring population
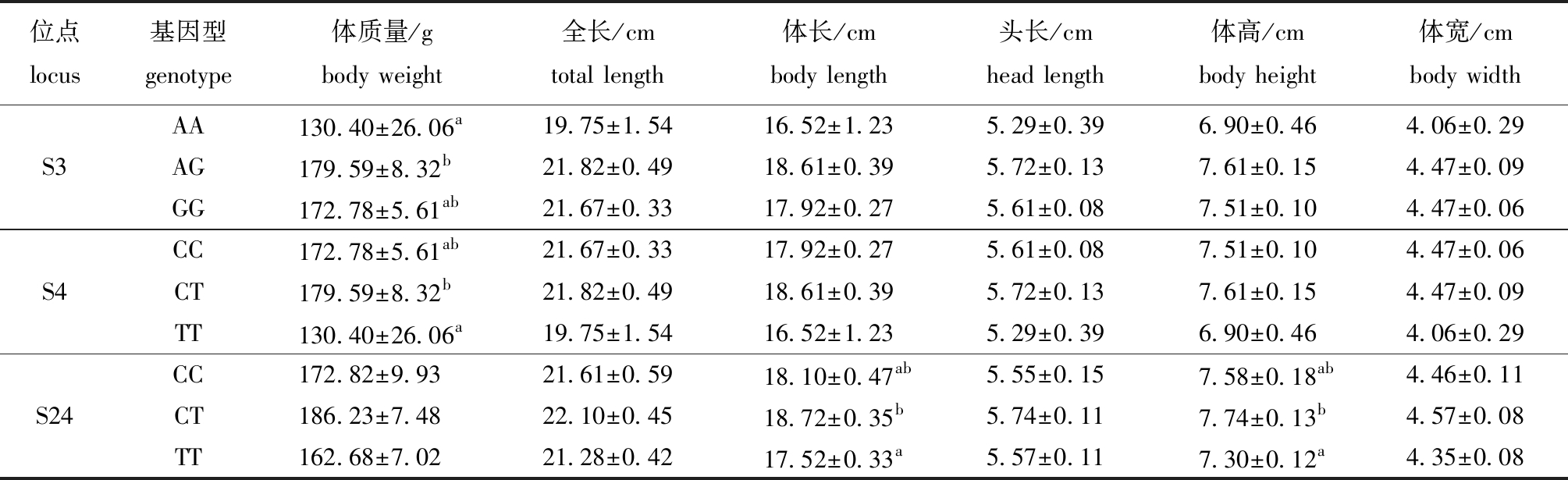
位点locus基因型genotype体质量/gbody weight全长/cmtotal length体长/cmbody length头长/cmhead length体高/cmbody height体宽/cmbody widthAA130.40±26.06a19.75±1.5416.52±1.235.29±0.396.90±0.464.06±0.29S3AG179.59±8.32b21.82±0.4918.61±0.395.72±0.137.61±0.154.47±0.09GG172.78±5.61ab21.67±0.3317.92±0.275.61±0.087.51±0.104.47±0.06CC172.78±5.61ab21.67±0.3317.92±0.275.61±0.087.51±0.104.47±0.06S4CT179.59±8.32b21.82±0.4918.61±0.395.72±0.137.61±0.154.47±0.09TT130.40±26.06a19.75±1.5416.52±1.235.29±0.396.90±0.464.06±0.29CC172.82±9.9321.61±0.5918.10±0.47ab5.55±0.157.58±0.18ab4.46±0.11S24CT186.23±7.4822.10±0.4518.72±0.35b5.74±0.117.74±0.13b4.57±0.08TT162.68±7.0221.28±0.4217.52±0.33a5.57±0.117.30±0.12a4.35±0.08
2.3.3 MYF5基因SNP位点与番禺群体生长性状的关联分析 从表8可见:S7位点AA基因型个体的体质量和全长均显著大于AT基因型个体(P<0.05);S21位点CC基因型个体的体质量显著大于CG、GG基因型个体(P<0.05);S32位点CC基因型个体的体质量显著大于CA基因型个体(P<0.05)。
表8 MYF5基因中各SNP位点与番禺群体生长性状的关联分析
Tab.8 Association analysis of SNP locus in MYF5 gene with growth traits of Panyu population
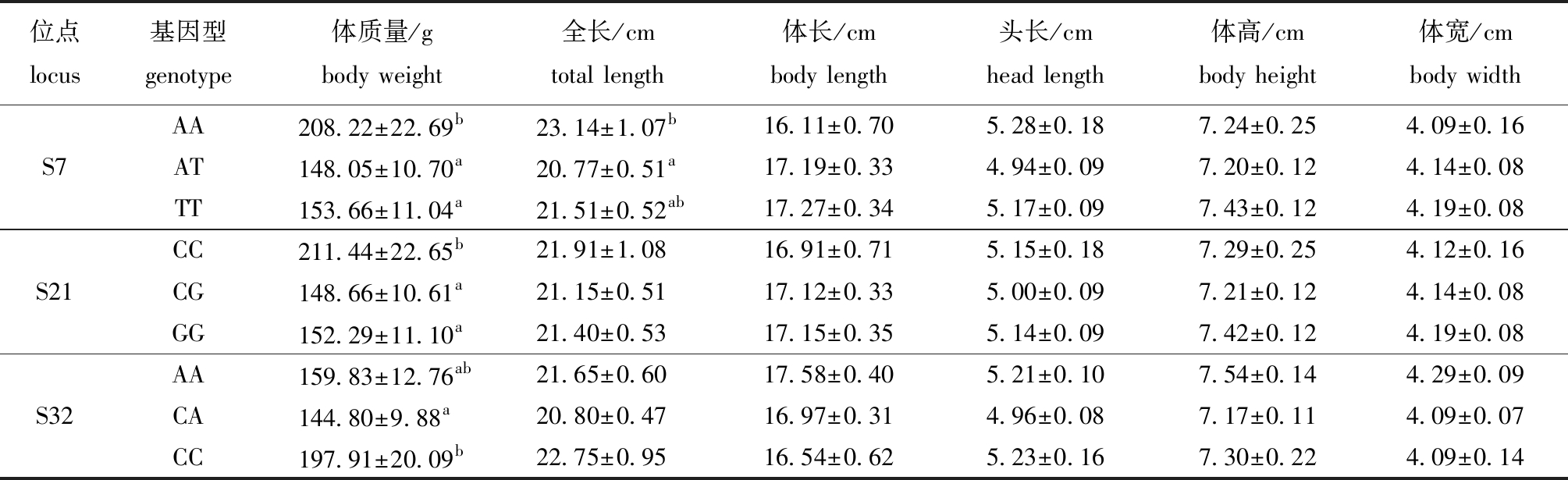
位点locus基因型genotype体质量/gbody weight全长/cmtotal length体长/cmbody length头长/cmhead length体高/cmbody height体宽/cmbody widthAA208.22±22.69b23.14±1.07b16.11±0.705.28±0.187.24±0.254.09±0.16S7AT148.05±10.70a20.77±0.51a17.19±0.334.94±0.097.20±0.124.14±0.08TT153.66±11.04a21.51±0.52ab17.27±0.345.17±0.097.43±0.124.19±0.08CC211.44±22.65b21.91±1.0816.91±0.715.15±0.187.29±0.254.12±0.16S21CG148.66±10.61a21.15±0.5117.12±0.335.00±0.097.21±0.124.14±0.08GG152.29±11.10a21.40±0.5317.15±0.355.14±0.097.42±0.124.19±0.08AA159.83±12.76ab21.65±0.6017.58±0.405.21±0.107.54±0.144.29±0.09S32CA144.80±9.88a20.80±0.4716.97±0.314.96±0.087.17±0.114.09±0.07CC197.91±20.09b22.75±0.9516.54±0.625.23±0.167.30±0.224.09±0.14
2.3.4 MYF5基因SNP位点与海南群体生长性状的关联分析 MYF5基因SNP位点与海南雌性、雄性群体的关联分析表明,均无与生长性状显著相关的SNP位点。
2.4 MYF5基因双倍型与各群体生长性状的相关性
2.4.1 MYF5基因双倍型与高要亲代群体生长性状的关联分析 将 29个SNP 位点进行单倍型分析,利用单倍型分析可能存在的双倍型(图2),去掉频率低于3% 的双倍型,将其余13种双倍型与生长性状进行关联分析,结果表明:D11与D13双倍型个体的全长显著大于D2双倍型个体(P<0.05);D22双倍型个体的头长显著大于D2双倍型个体(P<0.05);D18双倍型个体的体高显著大于D22双倍型个体(P<0.05)(表9)。
表9 MYF5基因双倍型与高要亲代群体生长性状的关联分析
Tab.9 Correlation analysis between growth traits and diplotype in the MYF5 gene in Gaoyao parent population
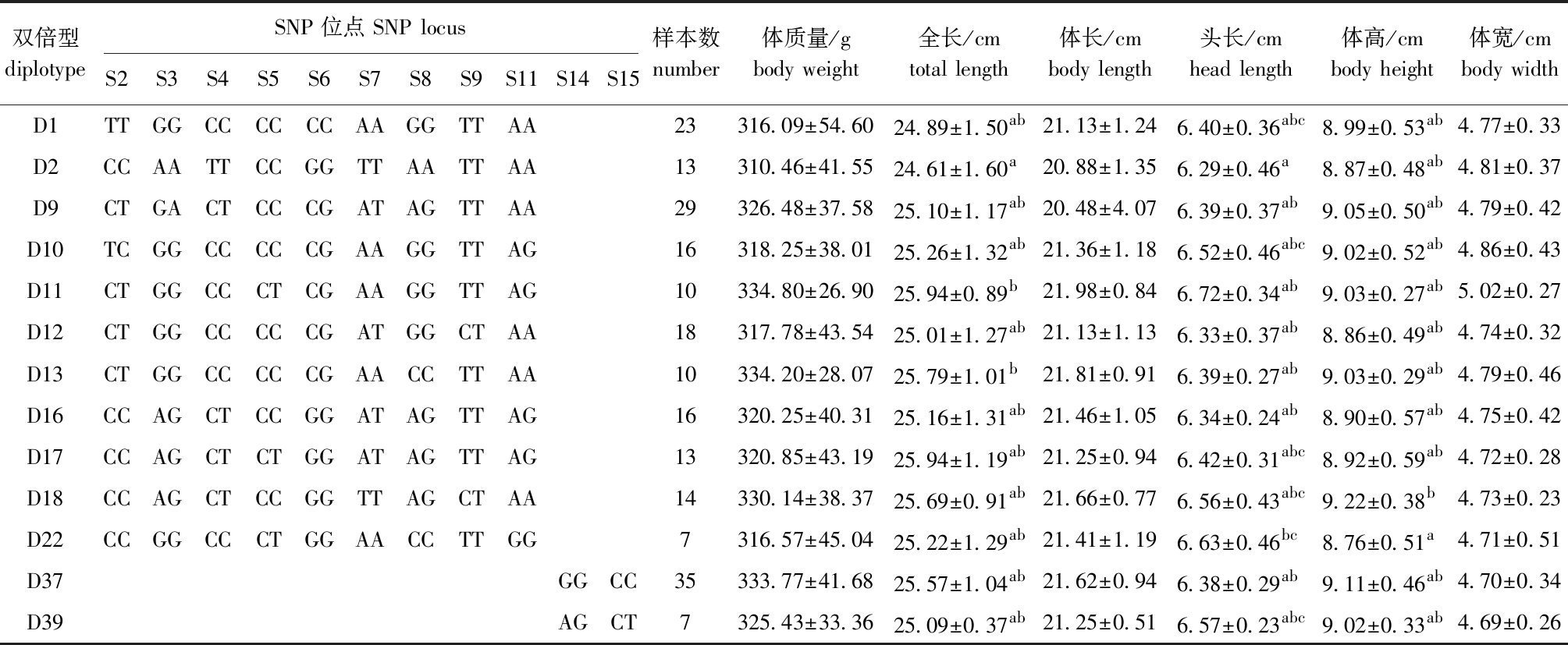
双倍型diplotypeSNP位点 SNP locusS2S3S4S5S6S7S8S9S11S14S15样本数number体质量/gbody weight全长/cmtotal length体长/cmbody length头长/cmhead length体高/cmbody height体宽/cmbody widthD1TTGGCCCCCCAAGGTTAA23316.09±54.6024.89±1.50ab21.13±1.246.40±0.36abc8.99±0.53ab4.77±0.33D2CCAATTCCGGTTAATTAA13310.46±41.5524.61±1.60a20.88±1.356.29±0.46a8.87±0.48ab4.81±0.37D9CTGACTCCCGATAGTTAA29326.48±37.5825.10±1.17ab20.48±4.076.39±0.37ab9.05±0.50ab4.79±0.42D10TCGGCCCCCGAAGGTTAG16318.25±38.0125.26±1.32ab21.36±1.186.52±0.46abc9.02±0.52ab4.86±0.43D11CTGGCCCTCGAAGGTTAG10334.80±26.9025.94±0.89b21.98±0.846.72±0.34ab9.03±0.27ab5.02±0.27D12CTGGCCCCCGATGGCTAA18317.78±43.5425.01±1.27ab21.13±1.136.33±0.37ab8.86±0.49ab4.74±0.32D13CTGGCCCCCGAACCTTAA10334.20±28.0725.79±1.01b21.81±0.916.39±0.27ab9.03±0.29ab4.79±0.46D16CCAGCTCCGGATAGTTAG16320.25±40.3125.16±1.31ab21.46±1.056.34±0.24ab8.90±0.57ab4.75±0.42D17CCAGCTCTGGATAGTTAG13320.85±43.1925.94±1.19ab21.25±0.946.42±0.31abc8.92±0.59ab4.72±0.28D18CCAGCTCCGGTTAGCTAA14330.14±38.3725.69±0.91ab21.66±0.776.56±0.43abc9.22±0.38b4.73±0.23D22CCGGCCCTGGAACCTTGG7316.57±45.0425.22±1.29ab21.41±1.196.63±0.46bc8.76±0.51a4.71±0.51D37GGCC35333.77±41.6825.57±1.04ab21.62±0.946.38±0.29ab9.11±0.46ab4.70±0.34D39AGCT7325.43±33.3625.09±0.37ab21.25±0.516.57±0.23abc9.02±0.33ab4.69±0.26
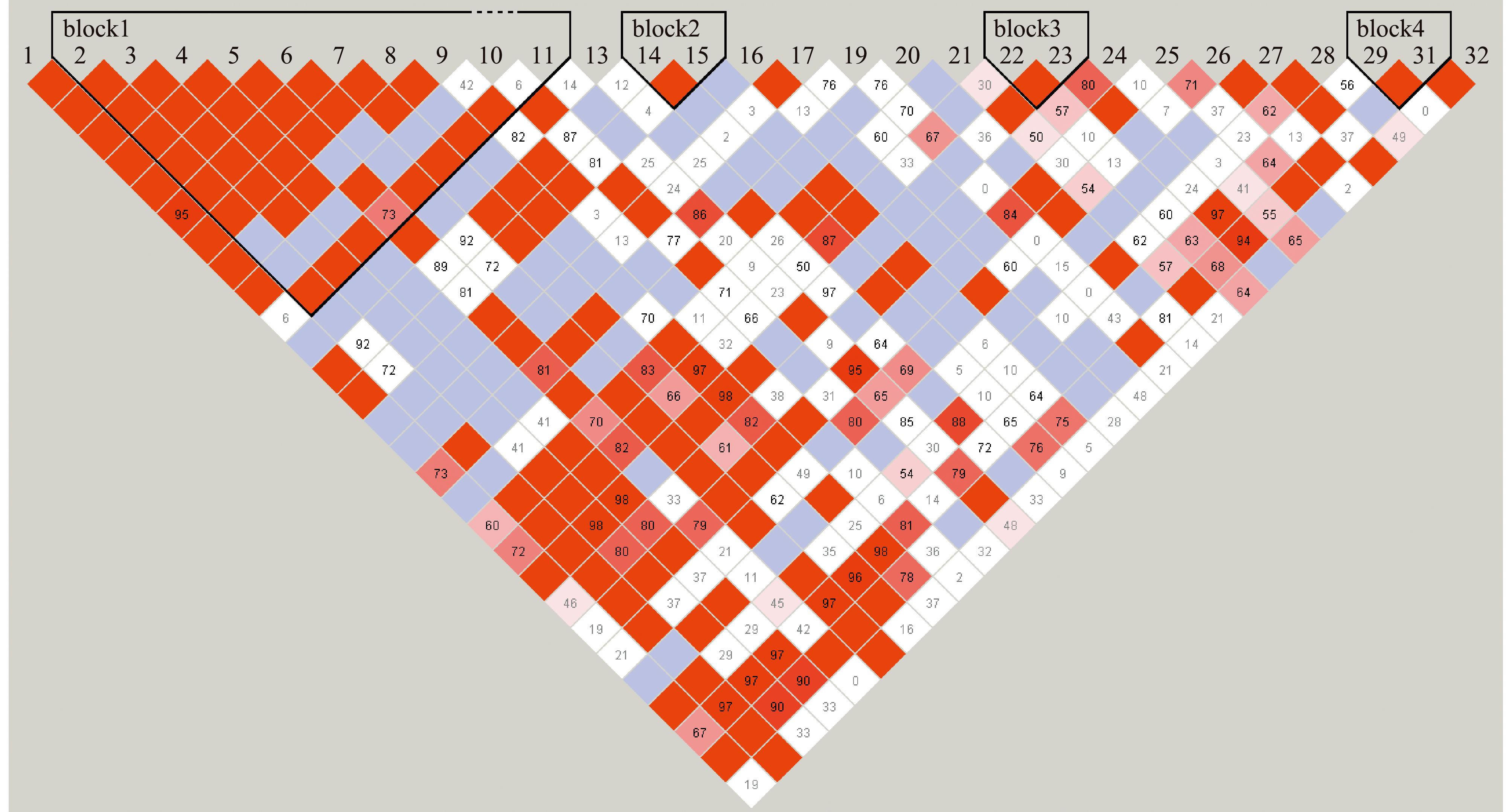
标记之间显示配对连锁不平衡(LD)值(r2),该值表示r2值乘以100;白色表示D=0,红色和粉色表示0<D<1,蓝色表示D=1,下同。
The values inside the squares indicate the linkage disequilibrium(LD)values(r2)of the two SNP locus. The value indicates that the r2 value is multiplied by 100. The color scheme is:white(D=0),red and pink(0<D<1)and blue(D=1),et sequentia.
图2 MYF5基因SNP位点在高要亲代群体中的连锁不平衡分析
Fig.2 Map of pair-wise LD between MYF5 SNP locus in Gaoyao parent populations
2.4.2 MYF5基因双倍型与高要子代群体生长性状的关联分析 将14个SNP位点进行单倍型分析,利用单倍型分析出双倍型(图3),去掉频率低于3%的双倍型,将其余11种双倍型与生长性状进行关联分析,结果表明,D11双倍型个体的体质量显著大于D3双倍型个体(P<0.05)(表10)。表明在实际育种应用中,应选择D11双倍型个体,以提高选育个体体质量。
表10 MYF5基因双倍型与高要子代群体生长性状的关联分析
Tab.10 Correlation analysis between growth traits and diplotype in the MYF5 gene in Gaoyao offspring population
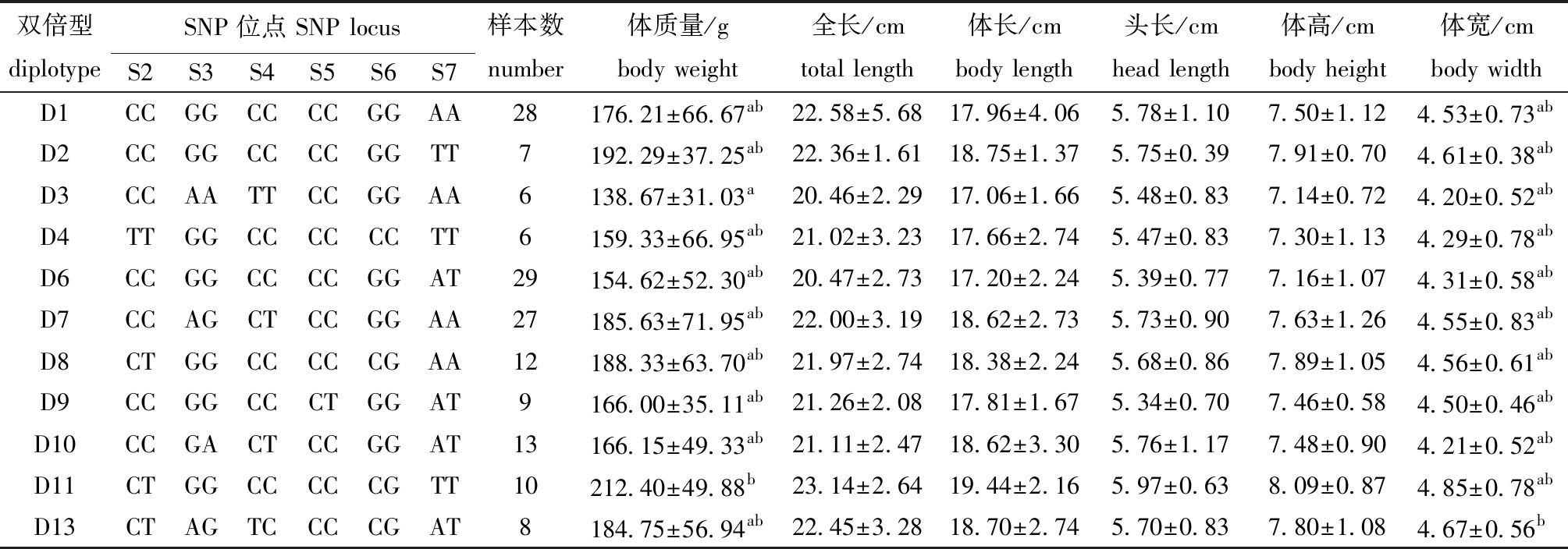
双倍型diplotypeSNP位点 SNP locusS2S3S4S5S6S7样本数number体质量/gbody weight全长/cmtotal length体长/cmbody length头长/cmhead length体高/cmbody height体宽/cmbody widthD1CCGGCCCCGGAA28176.21±66.67ab22.58±5.6817.96±4.065.78±1.107.50±1.124.53±0.73abD2CCGGCCCCGGTT7192.29±37.25ab22.36±1.6118.75±1.375.75±0.397.91±0.704.61±0.38abD3CCAATTCCGGAA6138.67±31.03a20.46±2.2917.06±1.665.48±0.837.14±0.724.20±0.52abD4TTGGCCCCCCTT6159.33±66.95ab21.02±3.2317.66±2.745.47±0.837.30±1.134.29±0.78abD6CCGGCCCCGGAT29154.62±52.30ab20.47±2.7317.20±2.245.39±0.777.16±1.074.31±0.58abD7CCAGCTCCGGAA27185.63±71.95ab22.00±3.1918.62±2.735.73±0.907.63±1.264.55±0.83abD8CTGGCCCCCGAA12188.33±63.70ab21.97±2.7418.38±2.245.68±0.867.89±1.054.56±0.61abD9CCGGCCCTGGAT9166.00±35.11ab21.26±2.0817.81±1.675.34±0.707.46±0.584.50±0.46abD10CCGACTCCGGAT13166.15±49.33ab21.11±2.4718.62±3.305.76±1.177.48±0.904.21±0.52abD11CTGGCCCCCGTT10212.40±49.88b23.14±2.6419.44±2.165.97±0.638.09±0.874.85±0.78abD13CTAGTCCCCGAT8184.75±56.94ab22.45±3.2818.70±2.745.70±0.837.80±1.084.67±0.56b
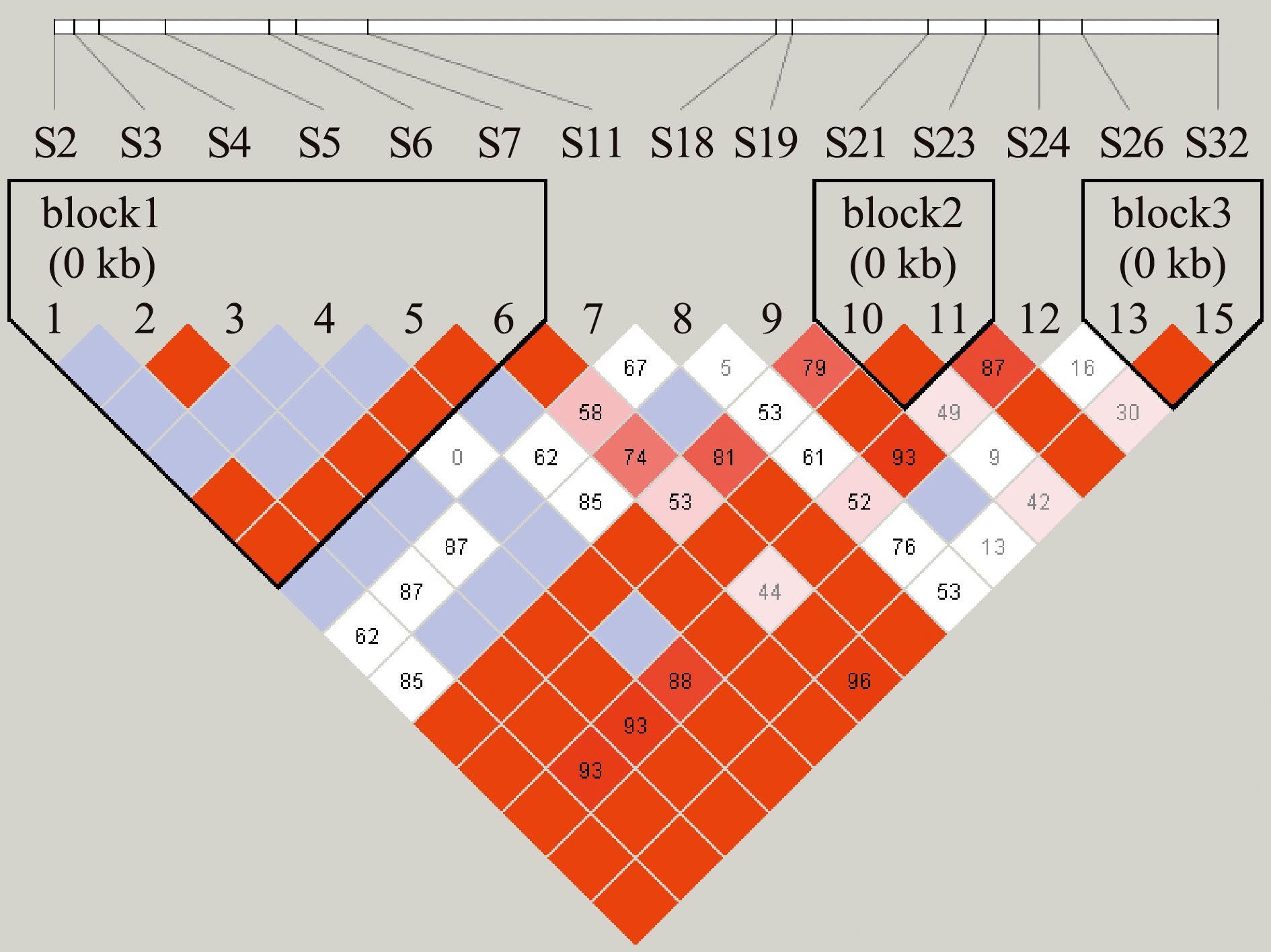
图3 MYF5基因SNP位点在高要子代群体中的连锁不平衡分析
Fig.3 Map of pair-wise LD between MYF5 SNP locus in Gaoyao offspring population
2.4.3 MYF5基因双倍型与番禺群体生长性状的关联分析 将14个SNP位点进行单倍型分析,利用单倍型分析出双倍型(图4),去掉频率低于3%的双倍型,将其余3种双倍型与生长性状进行关联分析,未发现与生长性状相关的双倍型(表11)。
表11 MYF5基因双倍型与番禺群体生长性状的关联分析
Tab.11 Correlation analysis between growth traits and diplotype in the MYF5 gene in Panyu population

双倍型diplotypeSNP位点 SNP locusS3S4样本数number体质量/gbody weight全长/cmtotal length体长/cmbody length头长/cmhead length体高/cmbody height体宽/cmbody widthD1GGCC121150.36±59.9421.18±4.1317.07±3.155.08±0.797.30±1.144.15±0.73D2AATT5158.80±68.7821.22±3.7917.72±3.174.89±0.887.33±1.234.06±0.89D3AGCT47173.19±160.2321.79±5.7417.18±2.645.10±0.767.33±0.874.18±0.53
图4 MYF5基因SNP位点在番禺群体中的连锁不平衡分析
Fig.4 Map of pair-wise LD between MYF5 SNP locus in Panyu population
2.4.4 MYF5基因双倍型与海南群体生长性状的关联分析 将9个SNP位点进行单倍型分析,利用单倍型分析出双倍型(图5),去掉频率低于3%的双倍型,将其余5种双倍型与海南雄性群体生长性状进行关联分析,未发现与生长性状相关的双倍型(表12)。海南雌性群体中,未发现形成单倍块(图6)。
表12 MYF5基因双倍型与海南雄性群体生长性状的关联分析
Tab.12 Correlation analysis between growth traits and diplotype in the MYF5 gene in Hainan male population
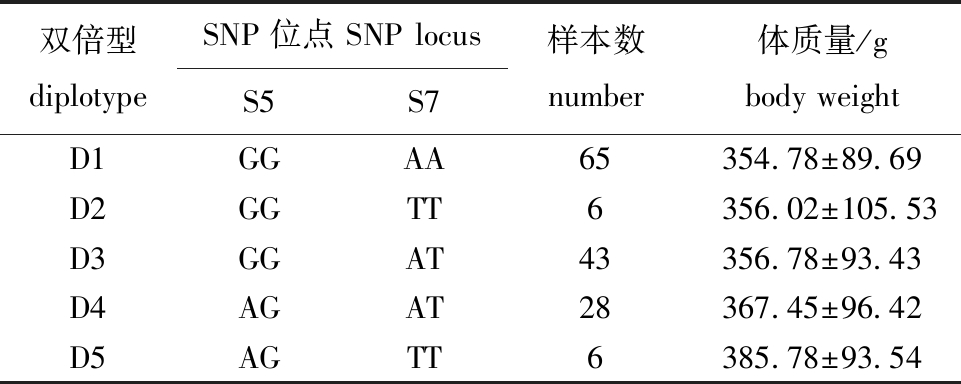
双倍型diplotypeSNP位点 SNP locusS5S7样本数number体质量/gbody weightD1GGAA65354.78±89.69D2GGTT6356.02±105.53D3GGAT43356.78±93.43D4AGAT28367.45±96.42D5AGTT6385.78±93.54
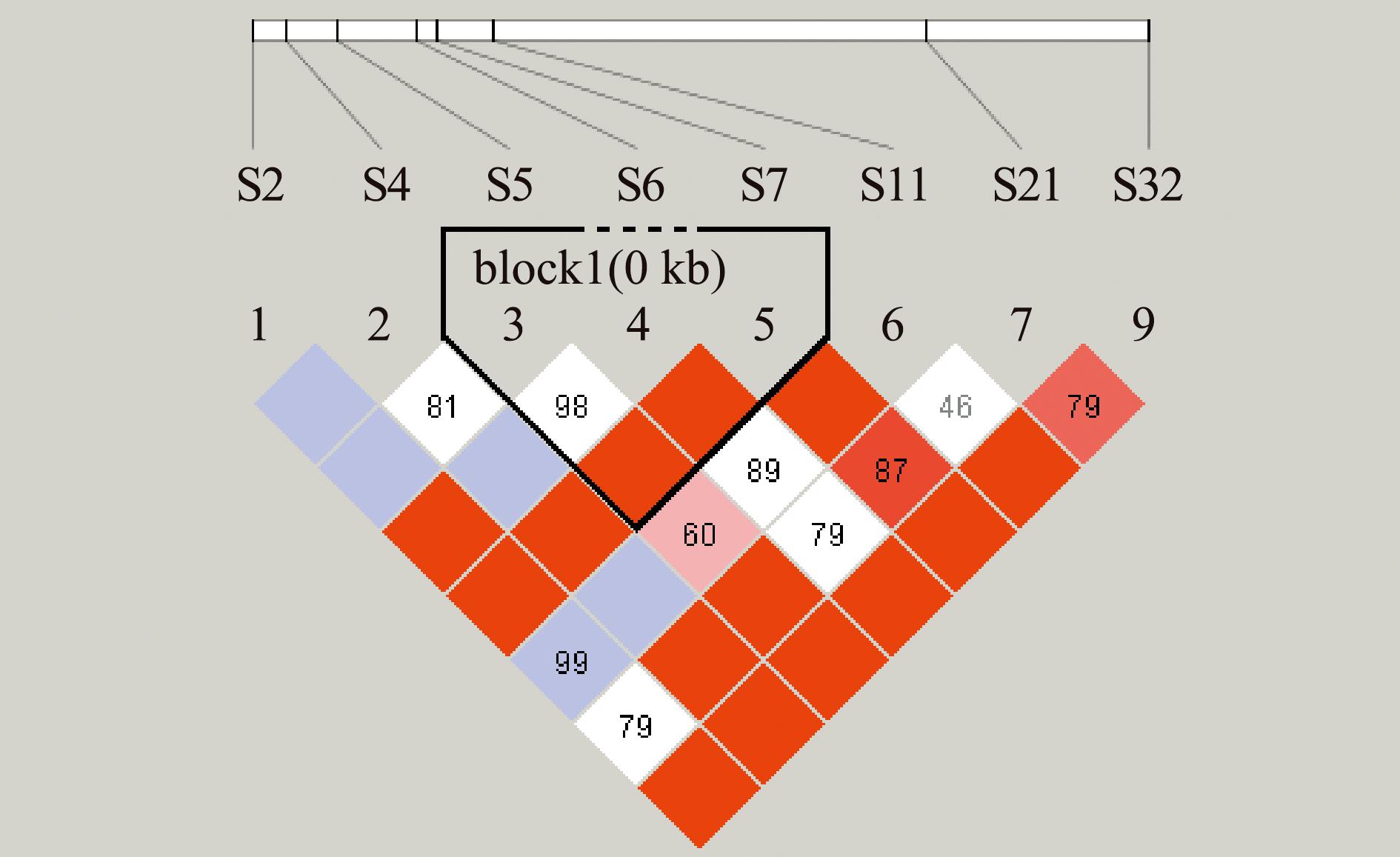
图5 MYF5 基因SNP位点在海南雄性群体中的连锁不平衡分析
Fig.5 Map of pair-wise LD between MYF5 SNP locus in Hainan male population
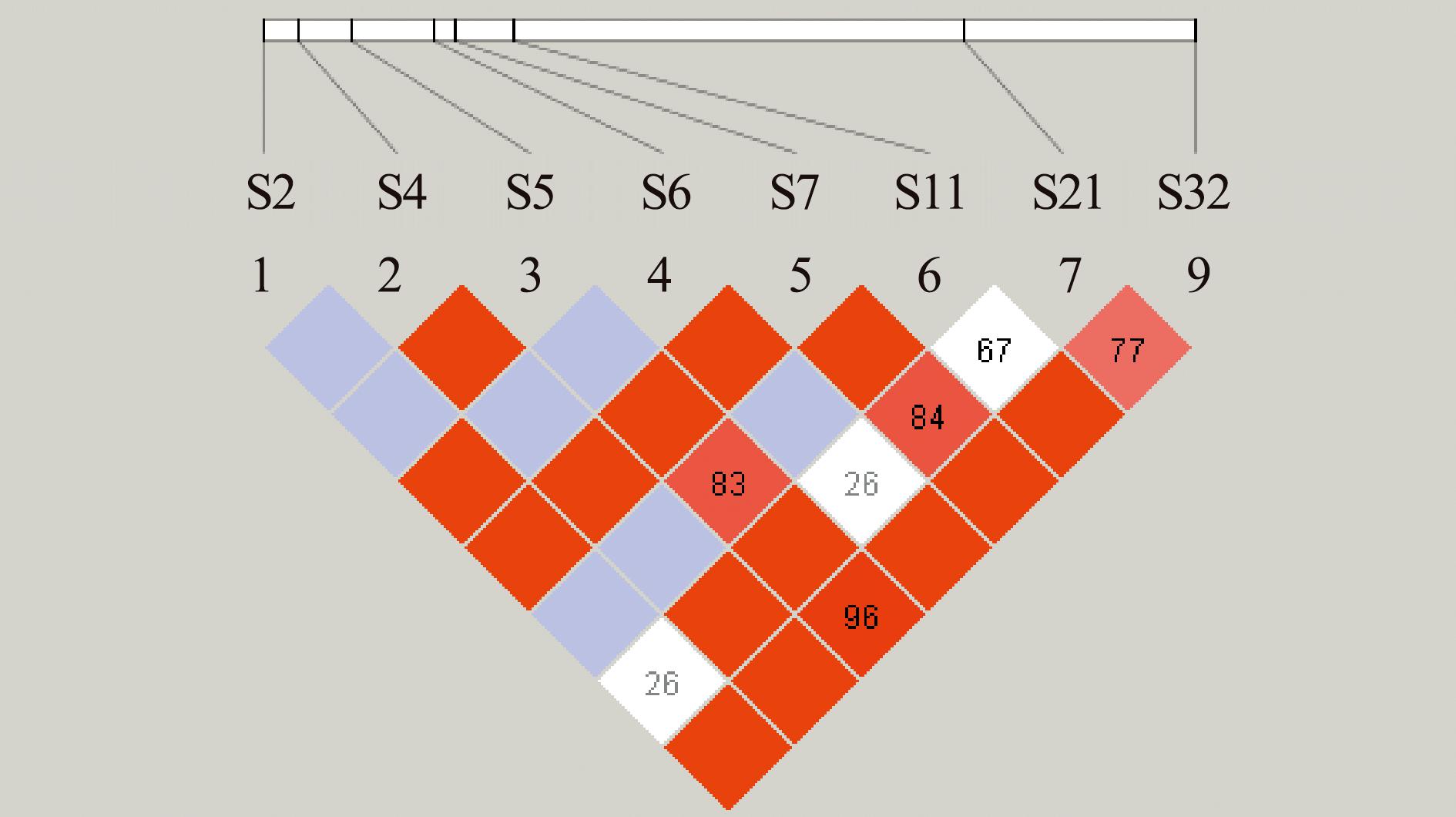
图6 MYF5 基因SNP位点在海南雌性群体中的连锁不平衡分析
Fig.6 Map of pair-wise LD between MYF5 SNP locus in Hainan female population
3 讨论
3.1 MYF5 SNP位点在尼罗罗非鱼群体中的分布
目前,对鱼类生长相关分子标记研究主要集中在生长轴相关基因和代谢相关基因[15-20],关于MyoD基因家族的研究主要集中在畜禽类。笔者对尼罗罗非鱼的研究表明,MYF5基因SNP在外显子1(550 bp)上有5个位点,平均密度为110 bp/位点,在外显子3(1 280 bp)上有16个位点,平均密度为80 bp/位点;在内含子2(223 bp)上有2个位点,平均密度为110 bp/位点,在内含子1(923 bp)上有11个位点,平均密度为84 bp/位点。内含子与外显子SNP位点密度基本一致,这与之前的研究结果不同,前期研究表明,内含子变异高于外显子[21-24]。景燕娟等[25]对大口黑鲈转录组研究显示,大口黑鲈基因编码区SNP位点密度为1/1 700 bp。本研究中,MYF5基因外显子SNP位点密度平均为95 bp/位点,远高于大口黑鲈基因编码区SNP位点密度,说明尼罗罗非鱼MYF5基因多态性较高,适合用于筛选与生长相关的SNP位点。
3.2 尼罗罗非鱼各群体的遗传多样性
等位基因数、杂合度和多态信息含量等遗传参数均能反映群体的遗传多样性水平,其数值的大小直接反映群体的基因丰富度和遗传多样性大小[26]。本研究中,高要亲代群体Ne、Ho两个多样性指标与子代群体相近,但He和PIC指标则低于子代群体。说明子代群体取样过程中可能样品代表性更好。高要子代群体平均PIC值为0.373 5,番禺群体平均PIC值为0.396 6,均稍高于王春晓等[18]2015年、2016年对这两个群体在GHSR基因、ghrelin基因中检测到的多样性。这可能是因为本研究所用的SNP位点数量及群体样本数量高于王春晓等[18]利用GHSR基因、ghrelin基因检测所用的位点及样本数量,故得出的数据更为准确。本研究中,获得的高要子代群体、番禺群体的PIC值均高于陈炳霖等[20]利用GHR、IGF-Ⅰ基因分析吉富罗非鱼群体及罗非鱼埃及群体遗传多样性的PIC值(分别为0.31和0.226)。Ho代表了群体遗传多样性。本研究表明,高要亲代群体、子代群体、番禺群体、海南雌群体和雄群体的Ho分别为0.263 3、0.267 6、0.324 5、0.259 3和0.274 8。Chen等[27]利用神经肽Y和食欲素前体基因SNP位点分析吉富罗非鱼群体和罗非鱼埃及群体遗传多样性,结果显示,两个群体的Ho分别为0.373 3和0.255。本研究中的各群体Ho与吉富罗非鱼群体和罗非鱼埃及群体相近。这表明,本研究中罗非鱼各群体均具有较高的遗传多样性,可以在此基础上继续进行良种选育。
本研究表明,MYF5基因在高要亲代群体中,S10、S13、S15、S16、S30均只有两种基因型。高要子代群体和番禺群体的S30位点只有一种基因型。这可能是选育过程中,由于人工选择,使得一些基因频率发生改变。如徐磊等[28]发现,大口黑鲈选育过程中,优势基因会进行富集。也有可能这些位点是不利位点,比如可能是致死位点。如Ma等[29]研究发现,大口黑鲈GHRH基因启动子上的缺失位点的BB基因型个体,在受精后全部死亡。
3.3 生长相关位点的筛选
鱼类生长特性直接影响鱼类产量,故鱼类生长特性在很大程度上决定了其经济回报。通过传统的育种方法来提高鱼类的生长性能是缓慢的。因此,许多研究都集中在鉴定生长性状的分子标记上。分子标记辅助选育,可以提高经济性状的选择效率。本研究中,从MYF5基因入手进行了与尼罗罗非鱼生长相关的SNP标记筛选,结果表明,MYF5基因外显子1上S3、S4位点与高要群体子代群体体质量显著相关。但这2个位点未引起编码氨基酸的改变。虽然该SNP可能与基因中具有功能意义区域的其他变异有关,但也可能直接影响基因功能。据报道,编码区中的同义SNP会影响mRNA的稳定性与翻译[30]、蛋白质折叠[31]、初级转录本的二级结构和交替剪接位点的出现[32]。内含子位点S18、S7分别与高要亲代群体、番禺群体生长性状显著关联。与外显子1中的同义SNP位点一样,内含子2 indel可能与基因中具有功能意义区域的其他变异有关。然而,内含子序列变化也可能直接影响基因表达,因为内含子会影响转录、多聚腺苷酸化、mRNA输出、翻译效率和mRNA衰减[33]。
3.4 生长相关SNP位点在各群体中的普适性
为了确保获得的与生长相关SNP位点的准确性,本研究中不仅将获得的与生长相关的位点在不同世代中进行验证,而且在不同群体中进行了验证。结果表明,在高要亲代群体中获得的生长相关位点,与在其子代中筛选到的生长相关位点有重合,但是与这些重合位点关联的生长性状有所不同。如S3和S4位点与高要亲代群体的体长显著相关,而与高要子代群体的体质量显著相关;S24位点与高要亲代群体的头长显著相关,而与高要子代群体的体长、体高均显著相关。这可能是亲代群体与子代群体生长状态不同导致的,也可能是因为鱼类生长性状是受多种基因控制的数量性状,存在主效基因和微效基因之分,且在不同世代中会出现分离或整合现象,从而导致筛选的生长相关SNP位点在不同世代之间适用性较差[34]。本研究中,在高要子代群体中获得的SNP位点与在番禺群体中获得的SNP位点不具有重复性。在高要子代群体中筛选到的生长相关位点为S3、S4、S24,而在番禺群体中筛选到的生长相关位点为S7、S21、S32。将这些位点在海南群体中进行进一步验证,未筛选到与海南群体生长相关的SNP位点。这可能是因为不同群体的遗传背景差异导致筛选到的生长相关SNP位点在不同群体中的关联性急剧下降。
4 结论
1)高要亲代群体中,尼罗罗非鱼MYF5基因S18位点与体质量和体宽显著相关,S3、S4位点与全长显著相关,S5和S24位点与头长显著相关。高要子代群体中,S3、S4位点与体质量显著相关,S24位点与体长、体高显著相关。这些位点可以作为候选基因标记,用于尼罗罗非鱼生长相关分子标记辅助育种研究。
2)将与高要子代群体及番禺群体生长性状相关的位点在尼罗罗非鱼海南群体中进行进一步验证,未获得与生长性状相关的位点。说明各生长相关位点在不同群体中普适性较低。
3)双倍型与各群体生长性状关联分析表明,在高要亲代群体中获得与全长、头长、体高显著相关的双倍型各1个,在高要子代群体中获得与体质量显著相关的双倍型1个,在番禺群体及海南群体中未获得与生长性状相关的双倍型。本研究结果可为改善尼罗罗非鱼生长性状提供有益参考。
[1] OLSON E N.MyoD family:a paradigm for development?[J].Genes &Development,1990,4(9):1454-1461.
[2] HUGHES S M,CHI M M,LOWRYO H,et al.Myogenin induces a shift of enzyme activity from glycolytic to oxidative metabolism in muscles of transgenic mice[J].The Journal of Cell Biology,1999,145(3):633-642.
[3] OTT M O,BOBER E,LYONS G,et al.Early expression of the myogenic regulatory gene,Myf-5,in precursor cells of skeletal muscle in the mouse embryo[J].Development,1991,111(4):1097-1107.
[4] TIMMONS J A,WENNMALM K,LARSSON O,et al.Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages[J].Proceedings of the National Academy of Sciences of the United States of America,2007,104(11):4401-4406.
[5] HOU G Y,ZHOU H L,CAO T,et al.Expression and variation of Myf5 and MyoD1 genes in different tissues of Wuzhishan pigs[J].Genetics and Molecular Research,2015,14(2):3729-3735.
[6] YAMAMOTO M,LEGENDREN P,BISWAS A A,et al.Loss of MyoD and Myf5 in skeletal muscle stem cells results in altered myogenic programming and failed regeneration[J].Stem Cell Reports,2018,10(3):956-969.
[7] ZHAO C P,RAZA S H A,KHAN R,et al.Genetic variants in MYF5 affected growth traits and beef quality traits in Chinese Qinchuan cattle[J].Genomics,2020,112(4):2804-2812.
[8] ZHANG Z C,LIU C,HAO W J,et al.Novelsingle nucleotide polymorphisms and haplotype of MYF5 gene are associated with body measurements and ultrasound traits in grassland short-tailed sheep[J].Genes,2022,13(3):483.
[9] TANG Y,ZHANG T,ZHANG G X,et al.Eight SNPs of the Myf5 gene and diplotypes associated with growth and reproductive traits in Jinghai yellow chicken[J].Molecular Biology Reports,2014,41(10):6837-6844.
[10] WANG J,HU Y S,ELZO M A,et al.Genetic effect of Myf5 gene in rabbit meat quality traits[J].Journal of Genetics,2017,96(4):673-679.
[11] 郭玉函,白俊杰,劳海华,等.大口黑鲈Myf5基因cDNA和基因组序列的克隆与分析[J].中国水产科学,2008,15(4):568-576.
GUO Y H,BAI J J,LAO H H,et al.Cloning and analysis of Myf5 cDNA and genomic sequence of largemouth bass (Micropterus salmoides)[J].Journal of Fishery Sciences of China,2008,15(4):568-576.(in Chinese)
[12] GUO Y H,BAI J J,CHANG O Q,et al.Molecular structure of the largemouth bass (Micropterus salmoides) Myf5 gene and its effect on skeletal muscle growth[J].Molecular Biology Reports,2009,36(6):1497-1504.
[13] 钟茂春,郑光明,赵建,等.鲮Myf5基因克隆及其SNPs分析[J].中国水产科学,2010,17(4):681-688.
ZHONG M C,ZHENG G M,ZHAO J,et al.Cloning and SNPs analysis of Myf5 gene from Cirrhinus molitorella[J].Journal of Fishery Sciences of China,2010,17(4):681-688.(in Chinese)
[14] CHEN B L,XIAO W,ZOU Z Y,et al.Ghrelin gene single nucleotide polymorphisms and their effects on Nile tilapia (Oreochromis niloticus) growth[J].Aquaculture Reports,2020,18:100469.
[15] 黄政举,何峰,温海深,等.半滑舌鳎IGF1基因编码区的多态性与生长性状及表达的相关性分析[J].中国海洋大学学报(自然科学版),2015,45(8):19-25.
HUANG Z J,HE F,WEN H S,et al.The polymorphism in the coding region of IGF1 related to mRNA expression pattern and growth traits of half-smooth tongue sole(Cynoglossus semilaevis)[J].Periodical of Ocean University of China(Natural Science Edition),2015,45(8):19-25.(in Chinese)
[16] 刘浩,白俊杰,李胜杰,等.大口黑鲈ghrelin基因SNPs的筛选及与生长性状关联性分析[J].水产学报,2016,40(4):521-527.
LIU H,BAI J J,LI S J,et al.SNPs detection of ghrelin gene and its association with growth traits in largemouth bass(Micropterus salmoides)[J].Journal of Fisheries of China,2016,40(4):521-527.(in Chinese)
[17] 刘浩,白俊杰,李胜杰,等.大口黑鲈PACAP基因SNPs的筛选及其与生长性状的关联分析[J].农业生物技术学报,2016,24(3):390-396.
LIU H,BAI J J,LI S J,et al.SNPs detection of PACAP gene and its association with growth traits in largemouth bass(Micropterus salmoides)[J].Journal of Agricultural Biotechnology,2016,24(3):390-396.(in Chinese)
[18] 王春晓,卢迈新,高风英,等.尼罗罗非鱼生长激素促分泌素受体基因(GHSR)生长相关单核苷酸多态性(SNPs)位点的筛选[J].农业生物技术学报,2015,23(6):762-771.
WANG C X,LU M X,GAO F Y,et al.Screening of single nucleotide polymophisms(SNPs) related with growth in growth hormone secretagogue receptor gene(GHSR) of Nile tilapia(Oreochromis niloticus)[J].Journal of Agricultural Biotechnology,2015,23(6):762-771.(in Chinese)
[19] 王春晓,卢迈新,高风英,等.尼罗罗非鱼ghrelin基因的多态性及其与生长性状相关SNP位点的筛选[J].水生生物学报,2016,40(1):50-57.
WANG C X,LU M X,GAO F Y,et al.The polymorphism of ghrelin gene of Oreochromis niloticus and identification of its SNP loci associated with the growth traits[J].Acta Hydrobiologica Sinica,2016,40(1):50-57.(in Chinese)
[20] 陈炳霖,肖炜,邹芝英,等.两种尼罗罗非鱼GHR、IGF-Ⅰ基因启动子区及编码区多态性与生长性状的相关性分析[J].农业生物技术学报,2020,28(11):2032-2047.
CHEN B L,XIAO W,ZOU Z Y,et al.Correlation analysis of polymorphisms in promoter region and coding region of GHR and IGF-Ⅰ genes with growth traits of two varieties of Nile tilapia(Oreochromis niloticus)[J].Journal of Agricultural Biotechnology,2020,28(11):2032-2047.(in Chinese)
[21] 陈雪峰,杨国梁,俞菊华,等.吉富罗非鱼IGF2基因分离及其单核苷酸多态性与体型、增重相关性[J].动物学杂志,2010,45(2):107-114.
CHEN X F,YANG G L,YU J H,et al.Isolation of IGF2 gene and correlation of its SNPs with fish sharp and weight gain in GIFT strain Nile tilapia Oreochromis niloticus[J].Chinese Journal of Zoology,2010,45(2):107-114.(in Chinese)
[22] HALUSHKA M K,FAN J B,BENTLEY K,et al.Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis[J].Nature Genetics,1999,22(3):239-247.
[23] RAFALSKI J A.Novel genetic mapping tools in plants:SNPs and LD-based approaches[J].Plant Science,2002,162(3):329-333.
[24] NIE Q H,LEI M M,OUYANG J H,et al.Identification and characterization of single nucleotide polymorphisms in 12 chicken growth-correlated genes by denaturing high performance liquid chromatography[J].Genetics,Selection,Evolution,2005,37(3):339-360.
[25] 景燕娟,白俊杰,李胜杰,等.大口黑鲈两个亚种EST数据库分析[J].上海海洋大学学报,2012,21(6):945-950.
JING Y J,BAI J J,LI S J,et al.Analysis of EST database for two subspecies of largemouth bass[J].Journal of Shanghai Ocean University,2012,21(6):945-950.(in Chinese)
[26] 朱冰,樊佳佳,白俊杰,等.金草鱼与中国4个草鱼群体的微卫星多态性比较分析[J].南方水产科学,2017,13(2):51-58.
ZHU B,FAN J J,BAI J J,et al.Gold grass carp microsatellite polymorphism and its comparative analysis with four grass carp populations from China[J].South China Fisheries Science,2017,13(2):51-58(in Chinese)
[27] CHEN B L,XIAO W,ZOU Z Y,et al.The effects of single nucleotide polymorphisms in Neuropeptide Y and prepro-orexin on growth in Nile tilapia (Oreochromis niloticus)[J].Aquaculture,2021,543:736974.
[28] 徐磊,白俊杰,李胜杰,等.生长相关优势基因型在大口黑鲈‘优鲈1号’选育世代中的聚合[J].华南农业大学学报,2014,35(1):7-11.
XU L,BAI J J,LI S J,et al.Pyramiding of growth-related genotypes in generations of the largemouth bass,Micropterus salmoides,‘Youlu No.1’[J].Journal of South China Agricultural University,2014,35(1):7-11.(in Chinese)
[29] MA D M,HAN L Q,BAI J J,et al.A 66-bp deletion in growth hormone releasing hormone gene 5′-flanking region with largemouth bass recessive embryonic lethal[J].Animal Genetics,2014,45(3):421-426.
[30] DUAN J B,WAINWRIGHT M S,COMERON J M,et al.Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor[J].Human Molecular Genetics,2003,12(3):205-216.
[31] KIMCHI-SARFATY C,OH J M,KIM I W,et al.A “silent” polymorphism in the MDR1 gene changes substrate specificity[J].Science,2007,315(5811):525-528.
[32] SAPRA A K,ARAVA Y,KHANDELIA P,et al.Genome-wide analysis of pre-mRNA splicing:intron features govern the requirement for the second-step factor,Prp17 in Saccharomyces cerevisiae and Schizosaccharomyces pombe[J].The Journal of Biological Chemistry,2004,279(50):52437-52446.
[33] NOTT A,MEISLIN S H,MOORE M J.A quantitative analysis of intron effects on mammalian gene expression[J].RNA,2003,9(5):607-617.
[34] 谭新,童金苟.SNPs及其在水产动物遗传学与育种学研究中的应用[J].水生生物学报,2011,35(2):348-354.
TAN X,TONG J G.SNPs and their applications in studies on genetics and breeding of aquaculture animals[J].Acta Hydrobiologica Sinica,2011,35(2):348-354.(in Chinese)