尼罗罗非鱼(Oreochromis niloticus)是中国重要特色淡水鱼类养殖品种之一,2021年中国罗非鱼总产量达到166万t,居世界首位[1]。随着罗非鱼产业的不断发展,种质混杂、退化造成的生长速率减慢,以及个体之间生长差异较大等问题日益严重,制约了罗非鱼养殖业的可持续发展。因此,以优良生长性状为目标的遗传改良成为罗非鱼种业发展的重要方向。目前,中国已审定的罗非鱼新品种有15个,包括选育种5个、杂交种5个、引进种3个和其他种2个,其中,绝大多数是以生长性状作为选育性状,采用传统杂交、群体选育和家系选育技术培育而成,尽管传统的基于表型的生长选择较为有效,但其选育周期长、选育效率低,且易受环境因素的影响,无法满足人们对优良品种不断更新换代的迫切需求[2]。分子标记辅助选择(MAS)可将传统育种中的表型选择转换为基因型选择,不易受环境、性别和年龄的影响,可进行早期选种和定向选育,缩短选育周期,显著提高遗传增益[3-4]。
分子标记有多种类型,包括随机扩增多态性DNA(RAPD)、扩增片段长度多态性(AFLP)、单链构象多态性(SSCP)、限制性片段长度多态性(RFLP)、单核苷酸多态性(SNP)及简单序列重复序列(SSR)等[5]。简单序列重复序列又称为微卫星DNA ,是广泛分布于真核生物基因组中的串联重复序列,属于共显性遗传且符合孟德尔遗传,其具有分布广、多态性丰富、检测方便和DNA用量少等特点,广泛应用于遗传图谱构建、数量性状定位、亲权鉴定、遗传多样性分析和分子辅助遗传育种等研究[6]。目前,在罗非鱼中已利用微卫星分子标记对多种质量和数量性状进行了QTL(quantitative trait locus)定位,包括耐寒[7]、耐盐[8-9]、耐低氧[10]、耐氨氮[11]、抗病[12-13]、体色[14]、性别[15-16]和生长[17-18]等性状,在此基础上进一步筛选与相应性状紧密连锁的微卫星位点及其基因型是利用分子标记辅助育种工作的重要环节。Liu等[17]在罗非鱼基因组中发现了10个体质量相关QTL、7个全长相关QTL和8个体长相关QTL,分布于6个连锁群。刘福平等[18]筛选到8个尼罗罗非鱼主要生长性状相关微卫星标记,并有3个位点在外群中得到进一步验证。李建林等[3]发现,位点GM041、GM304和UNH860的AA基因型及GM519的DE基因型可作为吉富罗非鱼体质量相关生长标记,且与体质量相关的标记越多,体质量越大,生长优势越明显。虽然罗非鱼生长相关微卫星标记已有少量报道,但由于检测方法、样本量大小及筛选群体遗传背景等差异导致其适用范围有限,利用已有标记难以开展罗非鱼快长品系的分子标记辅助选育工作[19]。因此,筛选更多遗传稳定、适用范围广的生长相关微卫星分子标记对基于不同基础群体的分子标记辅助选育具有重要意义。
本研究中,从高要F0群体大样本中筛选罗非鱼生长相关微卫星标记,在高要F1群体中进一步筛选遗传稳定的标记,通过扩大群体范围并在番禺群体和海南群体中分别进行验证,以期获得适用性较广的生长相关分子标记,为后续尼罗罗非鱼快长品系的选育工作提供指导。
1 材料与方法
1.1 材料
试验采集珠江水产研究所水产良种基地的尼罗罗非鱼F0群体(高要F0群体)尾鳍样本213尾和F1代群体(高要F1群体)尾鳍样本162尾,采集广东省罗非鱼良种场的尼罗罗非鱼(番禺群体)尾鳍样本175尾,采集海南省水产科学院的尼罗罗非鱼(海南群体)尾鳍样本300尾。将各鳍条样本置于无水乙醇中于-20 ℃保存备用。
高要群体为保种资源群体,其群体数量大且遗传多样性丰富,高要F0群体用于多态性微卫星位点的筛选和生长相关微卫星标记的初筛,其子代(高要F1群体)用于生长相关微卫星标记的遗传稳定性分析;番禺群体和海南群体为选育群体,用于生长相关微卫星标记的外群验证。
1.2 方法
1.2.1 生长性状测量 采用电子天平测量罗非鱼样本的体质量,采用游标卡尺测量其全长、体长、头长、体高和体宽等生长指标。
1.2.2 基因组DNA提取 采用动物组织基因组DNA提取试剂盒(Tiangen)提取上述样本基因组DNA。采用10 g/L琼脂糖凝胶电泳检测基因组DNA质量,采用超微量分光光度计(OSE-260,Tiangen)检测DNA浓度,并用无菌双蒸水将其稀释至质量浓度为50 ng/μL,于-20 ℃下保存备用。
1.2.3 微卫星引物与基因分型 从刘福平等[18]报道的生长相关微卫星位点和Liu等[17] 报道的罗非鱼生长性状相关QTL主要分布的6个连锁群(LG1、LG3、LG7、LG10、LG13和LG19)中随机筛选29个多态性较高的微卫星位点,引物序列从GenBank数据库中获取,在不同引物上游5′端加上不同荧光标记后,由广州天一辉远基因科技有限公司合成,引物信息详见表1。利用上述DNA模板进行扩增,PCR反应体系(共10 μL)包括:2×Taq PCR Master Mix 5.0 μL,基因组DNA(50 ng/μL)1 μL,上、下游引物(浓度为10 μmol/L)各0.5 μL,ddH2O 3 μL。反应程序:95 ℃下预变性5 min;95 ℃下变性30 s,50~66 ℃下退火30 s,72 ℃下延伸30 s,共进行10个循环;95 ℃下变性30 s,50 ℃下退火30 s,72 ℃下延伸30 s,共进行22个循环;最后在72 ℃下再延伸10 min。混合PCR产物后,用3730xl DNA Analyzer(ABI)测序仪进行多重毛细管电泳检测分型。
表1 29个尼罗罗非鱼微卫星位点及其引物信息
Tab.1 Primer information of 29 microsatellite loci in Oreochromis niloticus

位点locus引物序列(5′-3′)primer sequence(5′-3′)重复序列repeat array产物大小/bpproduct size连锁群linkage group登录号GenBank number退火温度/℃annealing temperatureGM114F:HEX-GCCTATTGAGCCTTGTGAGR:GTTTAACCCCACTATCCCTCT(GT)15138^1487BV005315.152.9GM124F:HEX-GTATCTGGGCAGTCATTTTCTR:TCATATTCTGATGGAGTGGG(CA)11311^39019BV005320.152.7GM165F:FAM-CTCATCAGTTGTTGCCTTCATR:CGGTAGAGCACTTCCTGTG(TG)11240^2557BV005339.155.0GM213F:HEX-TTTTATTCTGACAGGCACAR:AAAATCAAAGTTTAACATCCC(TG)24109^1385BV005364.151.2GM287F:HEX-TAGTGCCATCTCCATAAACACR:GTCTCCGTCGCTTCATT(CA)21204^22019BV005397.152.8GM336F:FAM-GCACCAGGAAGCAACTCAR:CGGCTGTGGCTATAAATAACA(CA)34214^2477BV005410.154.3GM386F:HEX-CCTAACAAGCGTCTTTTCAGCR:TGGCAAACAGTCAGTCAGTG(AC)18144^1677BV005435.156.0GM423F:TAMRA-GAGCGACTGTAGGACACGATTR:TGATGCTAAGAATGGCTGAGA(TG)14125^13019BV005448.156.8GM462F:ROX-AGTTTCCTTTTACACTGCTCCR:TCCACGGGTTACTGAAGA(GT)14210^2247BV005458.153.4GM565F:HEX-AGCCTCTGTGTTCAGCTTGGR:GGTTGGATGCCTATGATGTG(CA)11183^1987BV005501.155.7GM632F:TAMRA-GCAGCTACTCATGTGAGACCR:TGGGAACCTCTGGTCTATGC(TG)24160^17710BV005529.152.4GM634F:FAM-CCCGACATTAAACTTTCAGCAR:CTGAAACATGACTGCAGGAG(GT)26215^23819BV005531.153.4GM642F:HEX-CAGATCGTAACACAACGCAAAR:CACCCAGGACTGCTCTGTCT(GT)14229^24219BV005537.157.3Omo032F:HEX-GGTAAGCGGTGCAAAGTTCAR:GTTGGCTAACCTCTGTTCACTCC(GAAAA)10318^34213GR695143.158.8Omo170F:HEX-AGGGCAGCACTCTTTCACAGATTTR:AGGAGCAGGCCGAAAGGACAAT(GT)10256^2687JX20492963.6Omo184F:FAM-GGGGGCATGATTTACTTCTGAGGTR:AAGGCTGGAGCACAGGGACTTT(CA)12193^-21913JX20494264.8Omo216F:HEX-CGCCAGCAGAGGGCTAAGGAGR:AAGCAGAGTTCAGCCTCGCAGACT(GT)16436^4473JX20496166.0Omo377F:HEX-CTGACCCAGTGCACCATCTTR:TGAATTTAGAGGCTGTTTTGACAT(CA)12318^33710JX20508357.8Omo391F:FAM-AGACATCTGTACGCTCTTTACGAAR:AGTGCTAGAGGGAAGGGGCTGTA(GAT)9305^31113GR699257.158.3S002F:HEX-TTTCTCCCCAGCTTGTGTAACTCTR:TTGGACCTGGATTGCTGTTGATTA(CA)22374^4091KJ81209461.5S1541F:FAM-GGACAGATTCCATCCACCAGTAAR:TTCAAGCACATACAAAGCCTACAA(GT)12295^3081KJ81210256.5UNH115F:FAM-ACCTTCATCTCGGTCAGR:TCAAGCAGCTGATTTTT(CG)7(CA)16137^1673G12268.150.4UNH130F:HEX-AGGAAGAATAGCATGTAGCAAGTAR:GTGTGATAAATAAAGAGGCAGAAA(AC)23184^22923G12283.155.7UNH851F:FAM-TGGCTGTCACATAATCTGGTGR:CCTATCGTCGGTGATTGGTC(TG)15138^1507G68188.156.5UNH896F:FAM-CCTCTGTCCCTCCATGTGTTR:AGCCTGGCTTTAGAGGCAAT(GT)14150^2017G68214.156.7UNH914F:HEX-CAGCTTGTGGAAAGAAATACCAR:CCACGCACTTGTGGAAAATA(AC)11192^2027G68226.157.5UNH943F:FAM-CTGTCCGCCTTAAAGACCTGR:GCGCTCCTGAGGTTACTGTT(CA)29143^16219G68244.157.5UNH974F:ROX-GCACGTCTGAGAGTGTGGAAR:CAGCTTTCACACCAGCCTAA(GT)27186^21417G68261.156.2UNH985F:HEX-GCGTCTTGATGCAGGATACAR:TCCCGACGAGCAACTGTTAT(AC)22142^1541G68266.156.7
1.2.4 微卫星位点的遗传多样性分析 采用GeneMarker软件分析样本各位点扩增产物的大小,确定其基因型(同一位点的分子量由小到大依次标记为A、B、C、…)。采用Popgen 3.2软件分析微卫星位点的等位基因数(allele number,Na)、有效等位基因数(effective allele number Ne)、Shannon信息指数(Shannon information index,I)、观测杂合度(observed heterozygosity,Ho)和期望杂合度(expected heterozygosity,He)。采用PIC-CALC软件计算各位点的多态信息含量(polymorphism information content,PIC)。
1.3 数据处理
采用SPSS 19.0软件中Shapiro-Willk方法检验各群体生长性状是否符合正态分布,对不符合正态分布的性状进行对数(lg)转换后再进行检验。
采用一般线性模型(general linear model,GLM)对罗非鱼体质量、全长、体长、头长、体高和体宽6个生长性状与29个微卫星位点的相关性进行最小二乘分析,获得显著相关位点后利用Duncan法对微卫星标记不同基因型对应的生长性状进行多重比较。在统计分析中,只统计样本观测值≥5的基因型。
2 结果与分析
2.1 生长性状分布
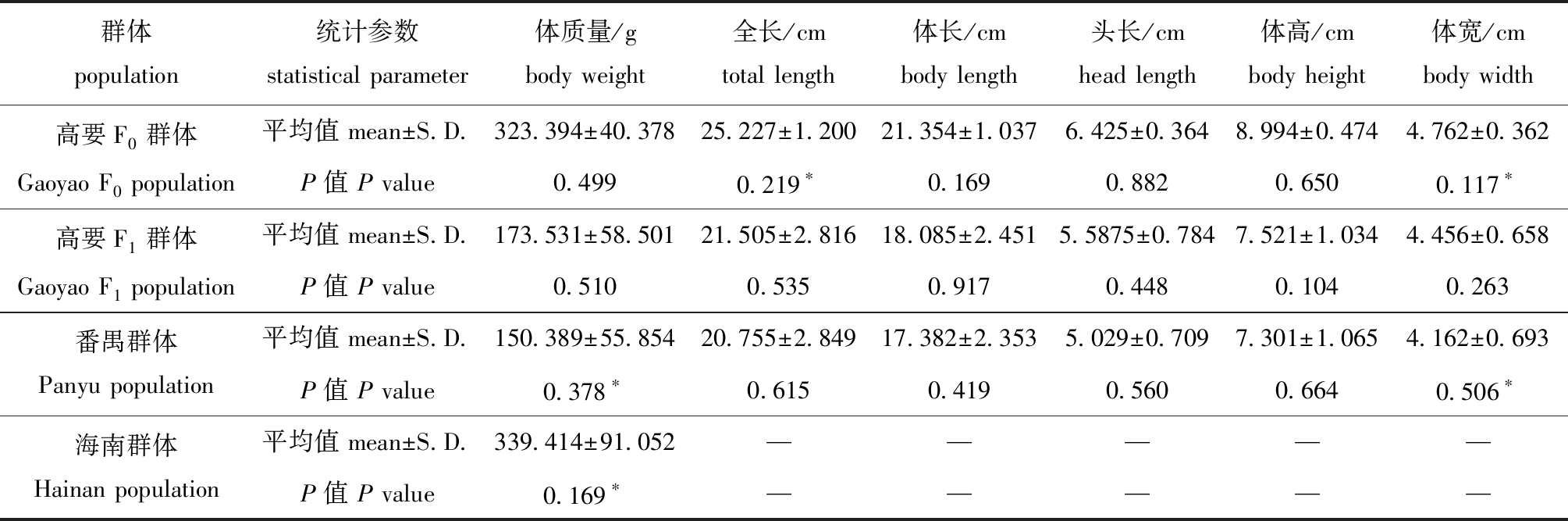
4个尼罗罗非鱼群体生长性状的平均值及其正态分布检验结果见表2,所有生长性状均表现为连续变化的特征,且绝大部分生长性状测量值符合正态分布(P>0.05),对不符合正态分布的生长性状测量值进行对数(lg)转换后也均符合正态分布。
表2 尼罗罗非鱼生长性状的正态分布检验
Tab.2 Normal distribution test of growth traits of Oreochromis niloticus
注:*表示生长性状数据经过对数(lg)转换之后的正态分布检验结果。
Note:*,indicates the normal distribution test result of growth traits after conversion by logarithmic(lg).
群体population统计参数statistical parameter体质量/gbody weight全长/cmtotal length体长/cmbody length头长/cmhead length体高/cmbody height体宽/cmbody width高要F0群体Gaoyao F0 population平均值 mean±S.D.323.394±40.37825.227±1.20021.354±1.0376.425±0.3648.994±0.4744.762±0.362P值 P value0.4990.219∗0.1690.8820.6500.117∗高要F1群体Gaoyao F1 population平均值 mean±S.D.173.531±58.50121.505±2.81618.085±2.4515.5875±0.7847.521±1.0344.456±0.658P值 P value0.5100.5350.9170.4480.1040.263番禺群体Panyu population平均值 mean±S.D.150.389±55.85420.755±2.84917.382±2.3535.029±0.7097.301±1.0654.162±0.693P值 P value0.378∗0.6150.4190.5600.6640.506∗海南群体Hainan population平均值 mean±S.D.339.414±91.052—————P值 P value0.169∗—————
2.2 微卫星标记的扩增及其多态性
29对微卫星引物在高要F0群体213个个体中能稳定扩增获得清晰条带,且多态性丰富、特异性强。共检测到242个等位基因,平均Na为8.345,平均Ne为3.973,平均I为1.540,平均Ho为0.621,平均He为0.710,平均PIC为0.842,各位点PIC值均大于0.5,具高度遗传多态性(表3)。
表3 尼罗罗非鱼高要F0群体中29个微卫星位点的遗传多样性参数
Tab.3 Genetic diversity parameters of 29 microsatellite loci in Gaoyao F0 population of Oreochromis niloticus
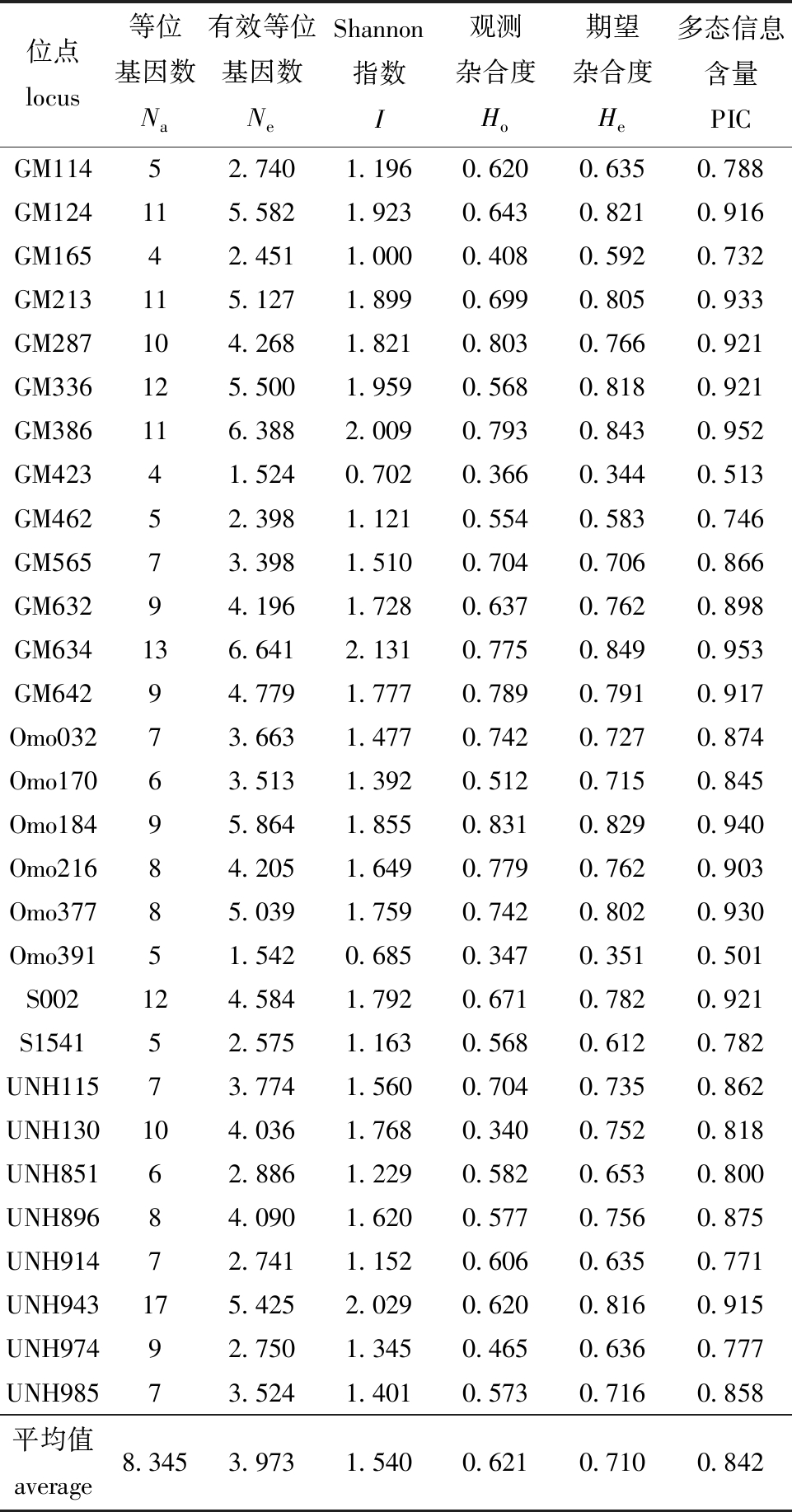
位点locus等位基因数Na有效等位基因数NeShannon指数I观测杂合度Ho期望杂合度He多态信息含量PICGM11452.7401.1960.6200.6350.788GM124115.5821.9230.6430.8210.916GM16542.4511.0000.4080.5920.732GM213115.1271.8990.6990.8050.933GM287104.2681.8210.8030.7660.921GM336125.5001.9590.5680.8180.921GM386116.3882.0090.7930.8430.952GM42341.5240.7020.3660.3440.513GM46252.3981.1210.5540.5830.746GM56573.3981.5100.7040.7060.866GM63294.1961.7280.6370.7620.898GM634136.6412.1310.7750.8490.953GM64294.7791.7770.7890.7910.917Omo03273.6631.4770.7420.7270.874Omo17063.5131.3920.5120.7150.845Omo18495.8641.8550.8310.8290.940Omo21684.2051.6490.7790.7620.903Omo37785.0391.7590.7420.8020.930Omo39151.5420.6850.3470.3510.501S002124.5841.7920.6710.7820.921S154152.5751.1630.5680.6120.782UNH11573.7741.5600.7040.7350.862UNH130104.0361.7680.3400.7520.818UNH85162.8861.2290.5820.6530.800UNH89684.0901.6200.5770.7560.875UNH91472.7411.1520.6060.6350.771UNH943175.4252.0290.6200.8160.915UNH97492.7501.3450.4650.6360.777UNH98573.5241.4010.5730.7160.858平均值 average8.3453.9731.5400.6210.7100.842
2.3 微卫星标记与高要F0群体生长性状的相关性
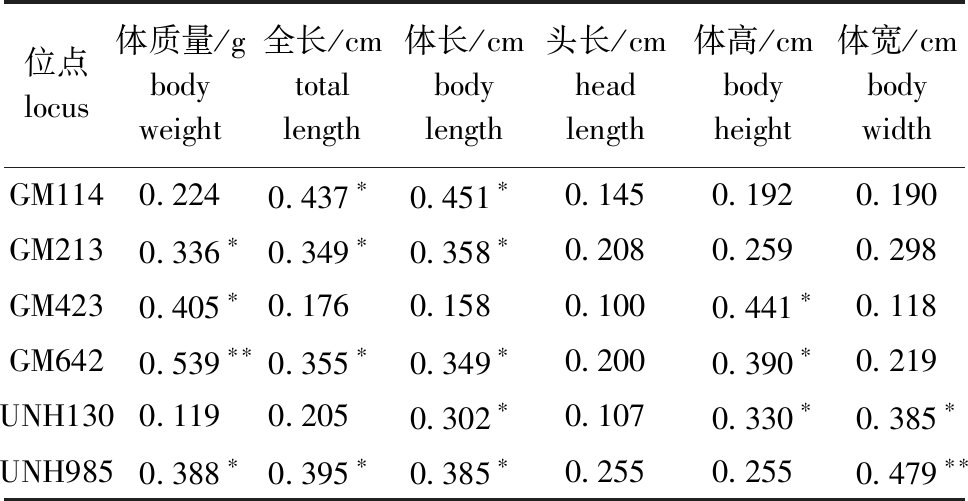
应用GLM模型对29个微卫星位点与尼罗罗非鱼高要F0群体生长性状的相关性进行最小二乘分析,结果显示,有6个微卫星位点与尼罗罗非鱼生长性状显著相关(表4)。其中,GM114位点与全长、体长显著相关(P<0.05);GM213位点与体质量、全长和体长显著相关(P<0.05);GM423位点与体质量、体高显著相关(P<0.05);GM642位点与体质量极显著相关(P<0.01),与全长、体长、体高显著相关(P<0.05);UNH130位点与体长、体高、体宽显著相关(P<0.05);UNH985位点与体质量、全长、体长显著相关(P<0.05),与体宽极显著相关(P<0.01)。
表4 微卫星位点与尼罗罗非鱼高要F0群体生长性状的相关系数(R)
Tab.4 Correlation coefficient(R)between microsatellite loci and growth traits of Gaoyao F0 population of Oreochromis niloticus
注:*表示显著相关(P<0.05);**表示极显著相关(P<0.01)。
Note:* means significant correlations(P<0.05);** means very significant correlations(P<0.01).
位点 locus体质量/g body weight全长/cm total length体长/cm body length头长/cm head length体高/cm body height体宽/cm body widthGM1140.2240.437∗0.451∗0.1450.1920.190GM2130.336∗0.349∗0.358∗0.2080.2590.298GM4230.405∗0.1760.1580.1000.441∗0.118GM6420.539∗∗0.355∗0.349∗0.2000.390∗0.219UNH1300.1190.2050.302∗0.1070.330∗0.385∗UNH9850.388∗0.395∗0.385∗0.2550.2550.479∗∗
2.4 微卫星标记与高要F1群体生长性状的相关性
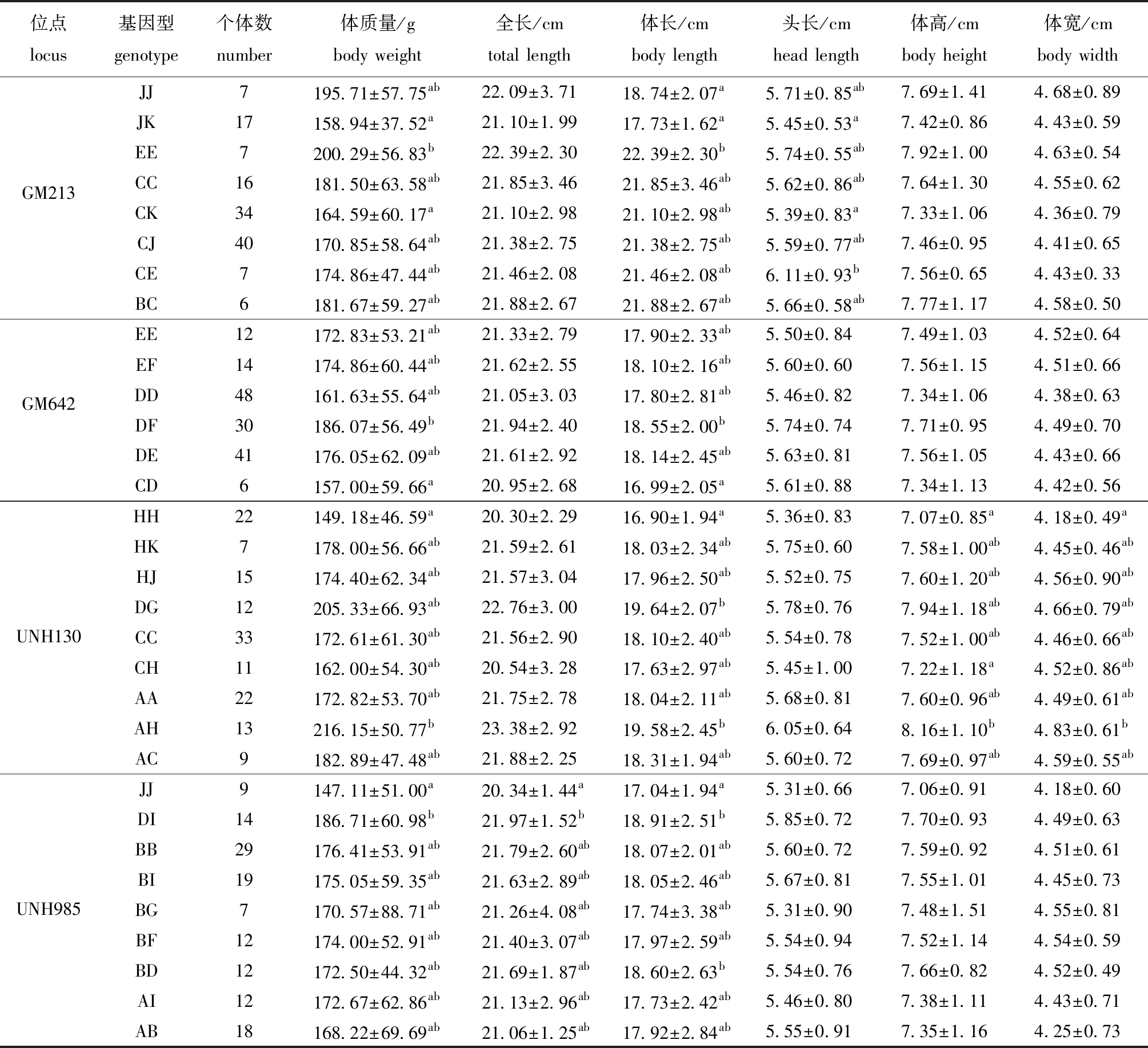
初筛的6个微卫星位点与尼罗罗非鱼高要F1群体生长性状的相关性分析显示,GM114和GM423位点不同基因型个体的生长性状之间无显著性差异(P>0.05),而GM213、GM642、UNH130和UNH985 4个位点的不同基因型个体的生长性状间存在显著性差异(P<0.05)(表5)。其中,GM213位点EE基因型个体的体质量显著高于JK和CK基因型(P<0.05),EE基因型个体的体长显著高于JJ和JK基因型(P<0.05),CE基因型个体的头长显著高于CK和JK基因型(P<0.05)。GM642位点DF基因型个体的体质量和体长显著高于CD基因型(P<0.05)。UNH130位点AH基因型个体的体质量和体宽均显著高于HH基因型(P<0.05),AH和DG基因型个体的体长显著高于HH基因型(P<0.05),AH基因型个体的体高显著高于CH和HH基因型(P<0.05)。UNH985位点DI基因型个体的体质量、全长和体长均显著高于JJ基因型(P<0.05)。
表5 高要F1群体中初筛微卫星位点不同基因型对应尼罗罗非鱼生长性状的平均值
Tab.5 Means of growth traits of different genotypes of preliminarily screened microsatellite locus in the Gaoyao F1 population of Oreochromis niloticus
注:同列中标有不同字母者表示同一微卫星位点相同性状的不同基因型间有显著性差异(P<0.05),标有相同字母者表示基因型间无显著性差异(P>0.05),下同。
Note:The means with different letters within the same microsatellite loci in same growth trait are significant differences in different genotypes at the P<0.05 probability level,and the means with the same letter are not significant differences,et sequentia.
位点locus基因型genotype个体数number体质量/gbody weight全长/cmtotal length体长/cmbody length头长/cmhead length体高/cmbody height体宽/cmbody widthGM213JJ7195.71±57.75ab22.09±3.7118.74±2.07a5.71±0.85ab7.69±1.414.68±0.89JK17158.94±37.52a21.10±1.9917.73±1.62a5.45±0.53a7.42±0.864.43±0.59EE7200.29±56.83b22.39±2.3022.39±2.30b5.74±0.55ab7.92±1.004.63±0.54CC16181.50±63.58ab21.85±3.4621.85±3.46ab5.62±0.86ab7.64±1.304.55±0.62CK34164.59±60.17a21.10±2.9821.10±2.98ab5.39±0.83a7.33±1.064.36±0.79CJ40170.85±58.64ab21.38±2.7521.38±2.75ab5.59±0.77ab7.46±0.954.41±0.65CE7174.86±47.44ab21.46±2.0821.46±2.08ab6.11±0.93b7.56±0.654.43±0.33BC6181.67±59.27ab21.88±2.6721.88±2.67ab5.66±0.58ab7.77±1.174.58±0.50GM642EE12172.83±53.21ab21.33±2.7917.90±2.33ab5.50±0.847.49±1.034.52±0.64EF14174.86±60.44ab21.62±2.5518.10±2.16ab5.60±0.607.56±1.154.51±0.66DD48161.63±55.64ab21.05±3.0317.80±2.81ab5.46±0.827.34±1.064.38±0.63DF30186.07±56.49b21.94±2.4018.55±2.00b5.74±0.747.71±0.954.49±0.70DE41176.05±62.09ab21.61±2.9218.14±2.45ab5.63±0.817.56±1.054.43±0.66CD6157.00±59.66a20.95±2.6816.99±2.05a5.61±0.887.34±1.134.42±0.56UNH130HH22149.18±46.59a20.30±2.2916.90±1.94a5.36±0.837.07±0.85a4.18±0.49aHK7178.00±56.66ab21.59±2.6118.03±2.34ab5.75±0.607.58±1.00ab4.45±0.46abHJ15174.40±62.34ab21.57±3.0417.96±2.50ab5.52±0.757.60±1.20ab4.56±0.90abDG12205.33±66.93ab22.76±3.0019.64±2.07b5.78±0.767.94±1.18ab4.66±0.79abCC33172.61±61.30ab21.56±2.9018.10±2.40ab5.54±0.787.52±1.00ab4.46±0.66abCH11162.00±54.30ab20.54±3.2817.63±2.97ab5.45±1.007.22±1.18a4.52±0.86abAA22172.82±53.70ab21.75±2.7818.04±2.11ab5.68±0.817.60±0.96ab4.49±0.61abAH13216.15±50.77b23.38±2.9219.58±2.45b6.05±0.648.16±1.10b4.83±0.61bAC9182.89±47.48ab21.88±2.2518.31±1.94ab5.60±0.727.69±0.97ab4.59±0.55abUNH985JJ9147.11±51.00a20.34±1.44a17.04±1.94a5.31±0.667.06±0.914.18±0.60DI14186.71±60.98b21.97±1.52b18.91±2.51b5.85±0.727.70±0.934.49±0.63BB29176.41±53.91ab21.79±2.60ab18.07±2.01ab5.60±0.727.59±0.924.51±0.61BI19175.05±59.35ab21.63±2.89ab18.05±2.46ab5.67±0.817.55±1.014.45±0.73BG7170.57±88.71ab21.26±4.08ab17.74±3.38ab5.31±0.907.48±1.514.55±0.81BF12174.00±52.91ab21.40±3.07ab17.97±2.59ab5.54±0.947.52±1.144.54±0.59BD12172.50±44.32ab21.69±1.87ab18.60±2.63b5.54±0.767.66±0.824.52±0.49AI12172.67±62.86ab21.13±2.96ab17.73±2.42ab5.46±0.807.38±1.114.43±0.71AB18168.22±69.69ab21.06±1.25ab17.92±2.84ab5.55±0.917.35±1.164.25±0.73
2.5 生长相关微卫星标记在外群中的验证
遗传稳定的4个生长相关微卫星位点(GM213、GM642、UNH130和UNH985)与番禺群体生长性状的相关性分析显示,4个位点各基因型个体的生长性状存在显著性差异(P<0.05)(表6)。其中,GM213位点的AA基因型个体的体质量显著高于CJ和JK基因型(P<0.05),BJ基因型个体的全长显著高于其他基因型(除AA型外)(P<0.05),BJ基因型个体的头长显著高于JK基因型(P<0.05)。GM642位点HH基因型个体的体质量显著高于DG和CD基因型(P<0.05),HH和BH基因型个体的体长显著大于DD、DG、CD和BD基因型(P<0.05),HH基因型个体的体高显著大于DG、CD和BD基因型(P<0.05)。UNH130位点FF基因型个体的体质量显著高于其他基因型(P<0.05)。UNH985位点CF基因型个体的体质量显著高于其他基因型(P<0.05),FK基因型的体长显著大于DK基因型(P<0.05)。
表6 番禺群体中4个微卫星位点不同基因型对应尼罗罗非鱼生长性状的平均值
Tab.6 Means of growth traits of different genotypes of 4 microsatellite loci in the Panyu population of Oreochromis niloticus
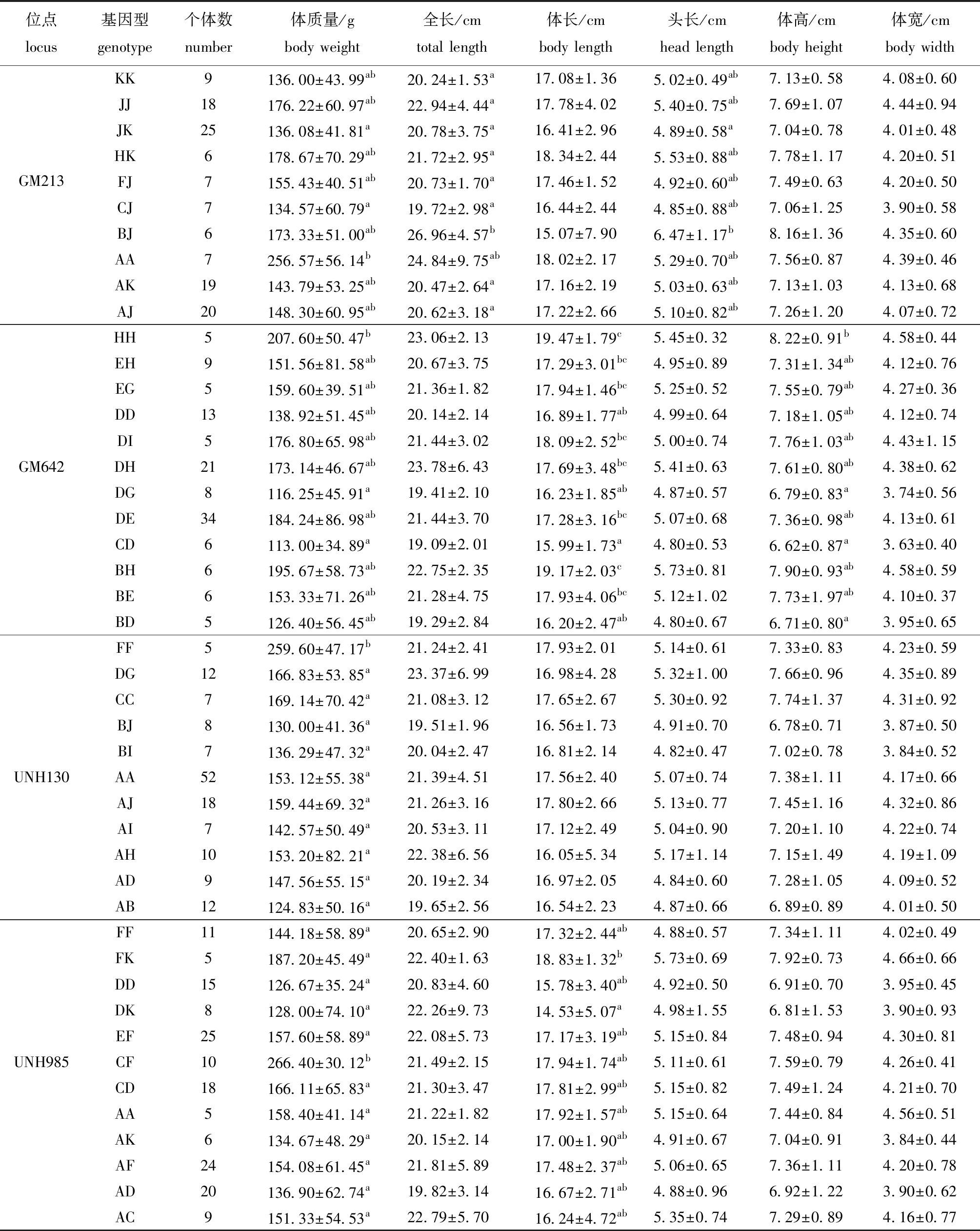
位点locus基因型genotype个体数number体质量/gbody weight全长/cmtotal length体长/cmbody length头长/cmhead length体高/cmbody height体宽/cmbody widthGM213KK9136.00±43.99ab20.24±1.53a17.08±1.365.02±0.49ab7.13±0.584.08±0.60JJ18176.22±60.97ab22.94±4.44a17.78±4.025.40±0.75ab7.69±1.074.44±0.94JK25136.08±41.81a20.78±3.75a16.41±2.964.89±0.58a7.04±0.784.01±0.48HK6178.67±70.29ab21.72±2.95a18.34±2.445.53±0.88ab7.78±1.174.20±0.51FJ7155.43±40.51ab20.73±1.70a17.46±1.524.92±0.60ab7.49±0.634.20±0.50CJ7134.57±60.79a19.72±2.98a16.44±2.444.85±0.88ab7.06±1.253.90±0.58BJ6173.33±51.00ab26.96±4.57b15.07±7.906.47±1.17b8.16±1.364.35±0.60AA7256.57±56.14b24.84±9.75ab18.02±2.175.29±0.70ab7.56±0.874.39±0.46 AK19143.79±53.25ab20.47±2.64a17.16±2.195.03±0.63ab7.13±1.034.13±0.68AJ20148.30±60.95ab20.62±3.18a17.22±2.665.10±0.82ab7.26±1.204.07±0.72GM642HH5207.60±50.47b23.06±2.1319.47±1.79c5.45±0.328.22±0.91b4.58±0.44EH9151.56±81.58ab20.67±3.7517.29±3.01bc4.95±0.897.31±1.34ab4.12±0.76EG5159.60±39.51ab21.36±1.8217.94±1.46bc5.25±0.527.55±0.79ab4.27±0.36DD13138.92±51.45ab20.14±2.1416.89±1.77ab4.99±0.647.18±1.05ab4.12±0.74DI5176.80±65.98ab21.44±3.0218.09±2.52bc5.00±0.747.76±1.03ab4.43±1.15DH21173.14±46.67ab23.78±6.4317.69±3.48bc5.41±0.637.61±0.80ab4.38±0.62DG8116.25±45.91a19.41±2.1016.23±1.85ab4.87±0.576.79±0.83a3.74±0.56DE34184.24±86.98ab21.44±3.7017.28±3.16bc5.07±0.687.36±0.98ab4.13±0.61CD6113.00±34.89a19.09±2.0115.99±1.73a4.80±0.536.62±0.87a3.63±0.40BH6195.67±58.73ab22.75±2.3519.17±2.03c5.73±0.817.90±0.93ab4.58±0.59BE6153.33±71.26ab21.28±4.7517.93±4.06bc5.12±1.027.73±1.97ab4.10±0.37BD5126.40±56.45ab19.29±2.8416.20±2.47ab4.80±0.676.71±0.80a3.95±0.65UNH130FF5259.60±47.17b21.24±2.4117.93±2.015.14±0.617.33±0.834.23±0.59DG12166.83±53.85a23.37±6.9916.98±4.285.32±1.007.66±0.964.35±0.89CC7169.14±70.42a21.08±3.1217.65±2.675.30±0.927.74±1.374.31±0.92BJ8130.00±41.36a19.51±1.9616.56±1.734.91±0.706.78±0.713.87±0.50BI7136.29±47.32a20.04±2.4716.81±2.144.82±0.477.02±0.783.84±0.52AA52153.12±55.38a21.39±4.5117.56±2.405.07±0.747.38±1.114.17±0.66AJ18159.44±69.32a21.26±3.1617.80±2.665.13±0.777.45±1.164.32±0.86AI7142.57±50.49a20.53±3.1117.12±2.495.04±0.907.20±1.104.22±0.74AH10153.20±82.21a22.38±6.5616.05±5.345.17±1.147.15±1.494.19±1.09AD9147.56±55.15a20.19±2.3416.97±2.054.84±0.607.28±1.054.09±0.52AB12124.83±50.16a19.65±2.5616.54±2.234.87±0.666.89±0.894.01±0.50UNH985FF11144.18±58.89a20.65±2.9017.32±2.44ab4.88±0.577.34±1.114.02±0.49FK5187.20±45.49a22.40±1.6318.83±1.32b5.73±0.697.92±0.734.66±0.66DD15126.67±35.24a20.83±4.6015.78±3.40ab4.92±0.506.91±0.703.95±0.45 DK8128.00±74.10a22.26±9.7314.53±5.07a4.98±1.556.81±1.533.90±0.93EF25157.60±58.89a22.08±5.7317.17±3.19ab5.15±0.847.48±0.944.30±0.81CF10266.40±30.12b21.49±2.1517.94±1.74ab5.11±0.617.59±0.794.26±0.41CD18166.11±65.83a21.30±3.4717.81±2.99ab5.15±0.827.49±1.244.21±0.70AA5158.40±41.14a21.22±1.8217.92±1.57ab5.15±0.647.44±0.844.56±0.51AK6134.67±48.29a20.15±2.1417.00±1.90ab4.91±0.677.04±0.913.84±0.44 AF24154.08±61.45a21.81±5.8917.48±2.37ab5.06±0.657.36±1.114.20±0.78AD20136.90±62.74a19.82±3.1416.67±2.71ab4.88±0.966.92±1.223.90±0.62AC9151.33±54.53a22.79±5.7016.24±4.72ab5.35±0.747.29±0.894.16±0.77
4个生长相关微卫星位点(GM213、GM642、UNH130和UNH985)与海南群体生长性状的相关性分析显示:GM213位点不同基因型个体的生长性状之间无显著性差异(P>0.05)(数据未列出);GM642位点DD基因型个体的体质量显著高于BB基因型(P<0.05);UNH130位点AD和BC基因型个体的体质量显著高于BG和FG基因型(P<0.05);UNH985位点GH基因型个体的体质量最大,DF基因型个体的体质量最小,二者间存在显著性差异(P<0.05)(表7)。
表7 海南群体中3个微卫星位点不同基因型对应尼罗罗非鱼体质量的平均值
Tab.7 Means of body weight of different genotypes of 3 microsatellite loci in the Hainan population of Oreochromis niloticus
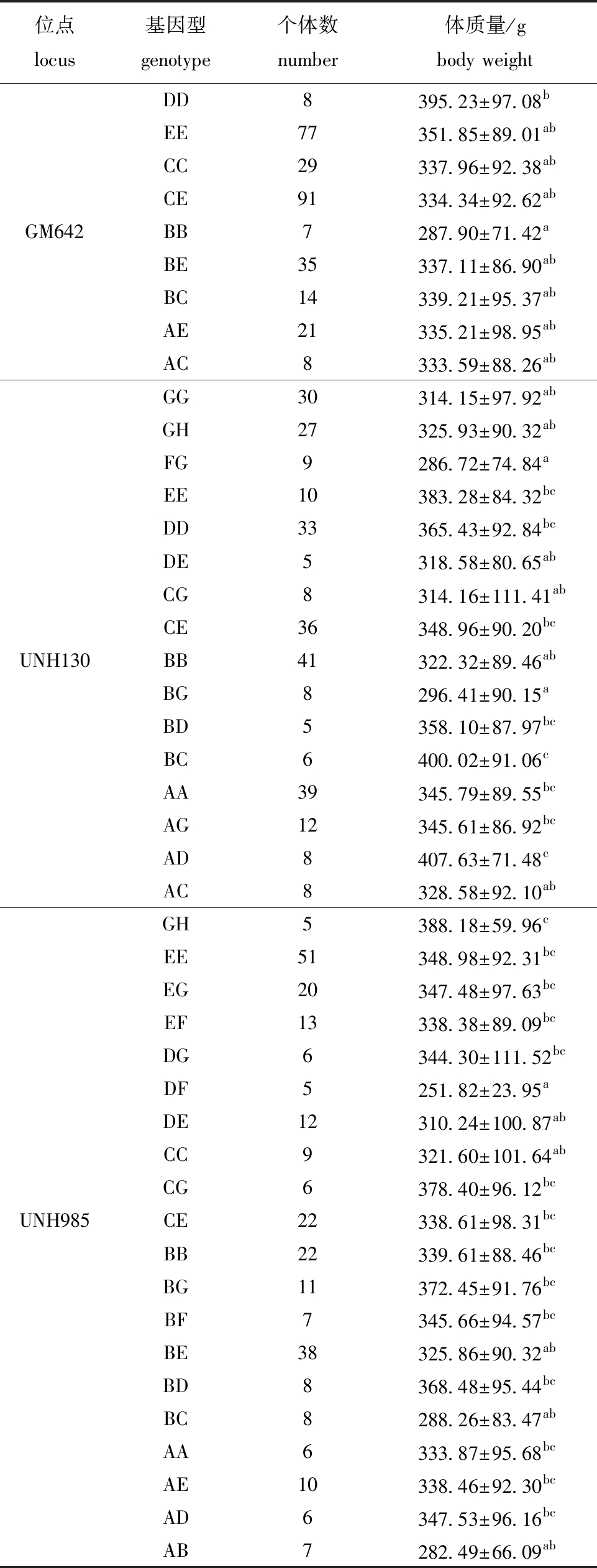
位点locus基因型genotype个体数number体质量/gbody weightDD8395.23±97.08bEE77351.85±89.01abCC29337.96±92.38abCE91334.34±92.62abGM642BB7287.90±71.42aBE35337.11±86.90abBC14339.21±95.37abAE21335.21±98.95abAC8333.59±88.26abGG30314.15±97.92abGH27325.93±90.32abFG9286.72±74.84aEE10383.28±84.32bcDD33365.43±92.84bcDE5318.58±80.65abCG8314.16±111.41abCE36348.96±90.20bcUNH130BB41322.32±89.46abBG8296.41±90.15aBD5358.10±87.97bcBC6400.02±91.06cAA39345.79±89.55bcAG12345.61±86.92bcAD8407.63±71.48cAC8328.58±92.10abGH5388.18±59.96cEE51348.98±92.31bcEG20347.48±97.63bcEF13338.38±89.09bcDG6344.30±111.52bcDF5251.82±23.95aDE12310.24±100.87abCC9321.60±101.64abCG6378.40±96.12bcUNH985CE22338.61±98.31bcBB22339.61±88.46bcBG11372.45±91.76bcBF7345.66±94.57bcBE38325.86±90.32abBD8368.48±95.44bcBC8288.26±83.47abAA6333.87±95.68bcAE10338.46±92.30bcAD6347.53±96.16bcAB7282.49±66.09ab
3 讨论
3.1 微卫星标记的遗传多样性
分子标记辅助育种是目前水产动物遗传育种的重要方向,它将分子遗传学与人工选择相结合,可在早期发育阶段进行选择,缩短了育种周期,同时可减少环境因素的影响,克服识别不明显性状的困难,从而显著提高选择的遗传响应和效率[20-21]。微卫星分子标记具有丰富的多态性,且在基因组中广泛分布,在水产动物分子辅助遗传育种中广泛应用。Barker[22]指出,至少具有4个等位基因的微卫星才能较好地用于遗传多样性分析。本研究中,采用的29个微卫星位点在高要F0群体中的等位基因数为4~17,平均等位基因数为8.345,满足上述要求,这与本研究中采用随机交配的外交群体遗传信息丰富、ABI 3730测序仪毛细管电泳分型方法精度较高,以及获得的等位基因数较多有关。DNA分子标记与数量性状之间关联的检测效率在很大程度上取决于资源群体中遗传变异的大小[23]。Botstein等[24]提出,当微卫星位点PIC<0.25时属于低度多态性水平,0.25≤PIC≤0.5时属于中度多态性水平,PIC>0.5时属于高度多态性水平。本研究中,29个微卫星位点在高要F0群体中的平均PIC为0.842,且各位点PIC值均大于0.5,均为高度多态性位点,这说明高要F0群体的杂合度较高,遗传多样性丰富,适用于分子标记的筛选。
3.2 微卫星标记与罗非鱼生长性状的关联分析
研究表明,当相关性系数为0~0.33时呈弱相关,相关系数为0.33~0.67时呈中度相关,相关系数为0.67~1.00时呈强相关[25]。本研究中,初筛获得6个微卫星标记在高要F0群体罗非鱼中与至少一种生长性状显著相关,其相关系数为0.302~0.539。说明这6个位点可能与罗非鱼生长性状相关主效基因连锁。也有研究表明,微卫星DNA可通过调节转录因子结合、DNA甲基化、启动子活性、增强子活性、mRNA稳定性、选择性剪接、核小体修饰和非编码RNA功能直接参与相关基因的表达调控[26-27]。本研究中,筛得的标记中存在一个标记与几种生长性状相关或几个标记与一种生长性状相关的现象,如UNH985位点与体质量、全长、体长和体宽均显著相关,体质量性状与GM213、GM423、GM642和UNH985位点均显著相关,说明这些位点的连锁基因存在一因多效和多因一效的现象,该现象在生长性状相关分子标记筛选的研究中较为常见,这与各生长性状间存在显著相关性及数量性状由多基因控制有关[28-29]。
在世代选育的过程中,分子标记在子代中的基因频率和基因型频率会发生改变,从亲本中筛得的生长相关标记的有效性也可能因此发生改变。本研究在高要F0群体中初筛的6个生长相关位点在其子代群体(高要F1群体)中的验证结果表明,有4个微卫星位点(GM213、GM642、UNH130和UNH985)的不同基因型个体间的生长指标具有显著性差异,表明这些标记在不同世代中遗传稳定。
遗传背景、环境因素及养殖方式等差异均会导致分子标记的应用范围存在局限性,在筛选群体中有效的分子标记可能在另一个群体中并不适用,因此,对分子标记在外群中进行验证是其应用的前提[30]。本研究中,在尼罗罗非鱼番禺群体和海南群体中对上述4个初筛生长相关微卫星位点的不同基因型进行多重比较,结果表明,番禺群体中4个微卫星标记(GM213、GM642、UNH130和UNH985)的不同基因型个体的生长性状之间存在显著差异;海南群体中有3个微卫星标记(GM642、UNH130和UNH985)的不同基因型个体的生长性状之间存在显著差异,这表明群体遗传背景差异对GM642、UNH130和UNH985 3个生长相关分子标记的影响较小,其适用范围较广,可作为首选的尼罗罗非鱼生长相关潜在分子标记。Liu等[17]在尼罗罗非鱼鹭业群体中发现了8个微卫星标记(UNH130、UNH183、UNH911、GM558、UNH211、UNH176、UNH914和UNH974)与主要生长性状显著或极显著相关,其中,UNH130在本研究中也被发现其与罗非鱼生长性状显著相关。
生长性状作为数量性状受多基因控制,利用一个或几个标记及其基因型无法将快长和慢长个体进行准确区分,在尼罗罗非鱼分子标记辅助选育工作中,将本研究中筛选的生长性状相关分子标记与已报道的相关分子标记联合使用,筛选富集更多生长相关优势基因型的个体,可获得更好的选育效果。
4 结论
1)GM642、UNH130和UNH985 3个位点在尼罗罗非鱼筛选群体和验证群体中均与生长性状显著相关,说明其受群体遗传背景差异的影响较小,适用范围较广,可作为重要候选生长性状相关分子标记。
2)GM642的DD和HH基因型,UNH130的AD、BC和FF基因型,以及UNH985的GH和CF基因型个体具有明显生长优势,说明这些基因型与优势生长性状连锁,在尼罗罗非鱼快长品系分子标记辅助育种中具有较好的应用潜力。
[1] 农业农村部渔业渔政管理局, 全国水产技术推广总站,中国水产学会.2022中国渔业统计年鉴[M].北京:中国农业出版社, 2022.
Fisheries and Fisheries Administration Bureau of Ministry of Agriculture and Rural Affairs,National Fisheries Technology Extension Center, China Society of Fisheries.China fisheries statistical yearbook 2022[M].Beijing:China Agriculture Press,2022.(in Chinese)
[2] LIN G,CHUA E,ORBAN L,et al.Mapping QTL for sex and growth traits in salt-tolerant tilapia (Oreochromis spp.×O.mossambicus)[J].PLoS One,2016,11(11):e0166723.
[3] 李建林,唐永凯,陈文华,等.吉富罗非鱼微卫星标记与体质量、体形性状相关性分析[J].中国水产科学,2009,16(6):824-832.
LI J L,TANG Y K,CHEN W H,et al.Association analysis of microsatellite DNA markers with body weight and body shape in GIFT[J].Journal of Fishery Sciences of China,2009,16(6):824-832.(in Chinese)
[4] EZE F.Marker-assisted selection in fish:a review[J].Asian Journal of Fisheries and Aquatic Research,2019,3(4):1-11.
[5] CHEN J L,FAN Z,TAN D J,et al.A review of genetic advances related to sex control and manipulation in tilapia[J].Journal of the World Aquaculture Society,2018,49(2):277-291.
[6] 刘志刚,曹建萌,高风英,等.罗非鱼“粤闽1号”母本选育群体世代间遗传差异的微卫星分析[J].大连海洋大学学报,2021,36(1):16-22.
LIU Z G,CAO J M,GAO F Y,et al.Genetic differentiation analysis of maternal selective breeding generations of tilapia “Yuemin No.1” using microsatellites[J].Journal of Dalian Ocean University,2021,36(1):16-22.(in Chinese)
[7] AI C H,LI B J,XIA J H.Mapping QTL for cold-tolerance trait in a GIFT-derived tilapia line by ddRAD-seq[J].Aquaculture,2022,556:738273.
[8] GU X H,JIANG D L,HUANG Y,et al.Identifying a major QTL associated with salinity tolerance in Nile tilapia using QTL-seq[J].Marine Biotechnology,2018,20(1):98-107.
[9] LIU F,SUN F,LI J,et al.A microsatellite-based linkage map of salt tolerant tilapia (Oreochromis mossambicus×Oreochromis spp.) and mapping of sex-determining loci[J].BMC Genomics,2013,14:58.
[10] LI H L,GU X H,LI B J,et al.Genome-wide QTL analysis identified significant associations between hypoxia tolerance and mutations in the GPR132 and ABCG4 genes in Nile tilapia[J].Marine Biotechnology,2017,19(5):441-453.
[11] ZHU Z X,LIN Y L,QIN H,et al.Identifying a genome-wide QTL interval controlling for ammonia-nitrogen tolerance on chrLG1 of Nile tilapia[J].Aquaculture,2021,543:736946.
[12] BARR A A,TRINH T Q,MAHMUDDIN M,et al.Correction to:a major quantitative trait locus affecting resistance to tilapia lake virus in farmed Nile tilapia (Oreochromis niloticus)[J].Heredity,2021,127(3):344.
A A,TRINH T Q,MAHMUDDIN M,et al.Correction to:a major quantitative trait locus affecting resistance to tilapia lake virus in farmed Nile tilapia (Oreochromis niloticus)[J].Heredity,2021,127(3):344.
[13] 朱佳杰,周宇,沈夏霜,等.吉富品系尼罗罗非鱼抗病性状分子标记筛选及遗传多样性分析[J].华北农学报,2013,28(sup 1):16-21.
ZHU J J,ZHOU Y,SHEN X S,et al.Screen of disease-resistance molecular markers and analysis of genetic diversity in GIFT strain Nile tilapia[J].Acta Agriculturae Boreali-Sinica,2013,28(sup 1):16-21.(in Chinese)
[14] LI B J,ZHU Z X,GU X H,et al.QTL mapping for red blotches in Malaysia red tilapia (Oreochromis spp.)[J].Marine Biotechnology,2019,21(3):384-395.
[15] ZHU Z X,LIN Y L,AI C H,et al.First identification of two co-existing genome-wide significant sex quantitative trait loci (QTL) in red tilapia using integrative QTL mapping[J].Zoological Research,2022,43(2):205-216.
[16] 陈文伟,曹建萌,刘志刚,等.罗非鱼性别相关微卫星标记的初步筛选[J].淡水渔业,2020,50(1):22-30.
CHEN W W,CAO J M,LIU Z G,et al.Preliminary screening of sex-related microsatellite markers in Oreochromis niloticus and O.aureus[J].Freshwater Fisheries,2020,50(1):22-30.(in Chinese)
[17] LIU F,SUN F,XIA J H,et al.A genome scan revealed significant associations of growth traits with a major QTL and GHR2 in tilapia[J].Scientific Reports,2014,4:7256.
[18] 刘福平,白俊杰,宋红梅,等.尼罗罗非鱼微卫星标记与主要生长性状的相关性分析[J].水产学报,2010,34(2):169-177.
LIU F P,BAI J J,SONG H M,et al.Correlation analysis of microsatellite DNA markers with major growth traits of tilapia(Oreochromis niloticus)[J].Journal of Fisheries of China,2010,34(2):169-177.(in Chinese)
[19] 鲁翠云,曹顶臣,李超,等.生长相关的微卫星标记在镜鲤繁殖群体中的验证[J].天津农学院学报,2012,19(3):13-18.
LU C Y,CAO D C,LI C,et al.Verification of microsatellite markers correlated with growth trait in mirror carp Cyprinus carpio L[J].Journal of Tianjin Agricultural University,2012,19(3):13-18.(in Chinese)
[20] LANDE R,THOMPSON R.Efficiency of marker-assisted selection in the improvement of quantitative traits[J].Genetics,1990,124(3):743-756.
[21] SONESSON A K.Within-family marker-assisted selection for aquaculture species[J].Genetics,Selection,Evolution,2007,39(3):301-317.
[22] BARKER J S F.A global protocol for determining genetic distances among domestic livestock breed[C]//Proceedings of the 5th world congress on genetics applied to livestock production.Canada:University of Guelph,1994:501-508.
[23] YUNIS R,HELLER E D,HILLEL J,et al.Microsatellite markers associated with quantitative trait loci controlling antibody response to Escherichia coli and Salmonella enteritidis in young broilers[J].Animal Genetics,2002,33(6):407-414.
[24] BOTSTEIN D,WHITE R L,SKOLNICK M,et al.Construction of a genetic linkage map in man using restriction fragment length polymorphisms[J].American Journal of Human Genetics,1980,32(3):314-331.
[25] 马爱军,邹杰,孙建华,等.暗纹东方鲀生长性状相关微卫星标记筛选[J].海洋科学,2016,40(10):16-24.
MA A J,ZOU J,SUN J H,et al.Screening growth-related microsatellite markers in Takifugu obscurus[J].Marine Sciences,2016,40(10):16-24.(in Chinese)
[26] LI Y C,KOROLAB,FAHIMA T,et al.Microsatellites within genes:structure,function,and evolution[J].Molecular Biology and Evolution,2004,21(6):991-1007.
[27] BAGSHAW A T M.Functional mechanisms of microsatellite DNA in eukaryotic genomes[J].Genome Biology and Evolution,2017,9(9):2428-2443.
[28] 李婷,李伟,赵建,等.中华鳖(Trionyx sinensis)微卫星标记与生长性状的相关分析[J].基因组学与应用生物学,2016,35(1):63-71.
LI T,LI W,ZHAO J,et al.Correlation analysis of the microsatellite DNA markers and growth traits of Chinese soft shell turtle(Trionyx sinensis)[J].Genomics and Applied Biology,2016,35(1):63-71.(in Chinese)
[29] 刘伟,苏胜彦,董在杰,等.3个鲤群体的微卫星标记与生长性状相关性分析[J].南方水产科学,2012,8(3):17-24.
LIU W,SU S Y,DONG Z J,et al.Correlation analysis of microsatellite DNA markers with growth trait among 3 breeding populations of common carp[J].South China Fisheries Science,2012,8(3):17-24.(in Chinese)
[30] HOU S Y,MA A J,WANG X A,et al.Isolation and characterization of 45 polymorphie microsatellite loci of turbot (Scophthalmus maximus) and cross-species amplification[J].Chinese Journal of Oceanology and Limnology,2011,29(2):311-316.