乌龟 (Mauremys reevesii),又名中华草龟、金龟,隶属于龟鳖目(Plestudinata)淡水龟科(Bataguridae)拟水龟属(Mauremys),是中国现存龟类中数量最多、分布最广和养殖数量最大的品种之一,年产量可达4万~5万t。乌龟全身都是宝,龟板是中国传统的珍贵药材,龟肉和龟血也都是高级滋补品和食疗佳品[1],其较高的营养、药用和观赏性获得了人们的广泛认可,市场需求旺盛,乌龟养殖前景广阔。
自20世纪80年代乌龟人工繁育技术开展以来,研究者在乌龟的基础与应用研究方面均取得了突破。随着乌龟养殖规模的快速扩大,种苗需求旺盛,而野生来源的亲本逐渐被消耗,人工培育的亲本成为乌龟养殖业的主要苗种来源[2]。然而,由于缺乏系统选育,各苗种场种质混杂和退化问题日益凸显,苗种供应仍存在着质量良莠不齐、病害多发、生产管理不规范及工厂化育苗模式不完善等诸多制约生产规模有效扩大的问题。因此,开展乌龟品种选育工作对于乌龟产业的可持续利用具有重要意义。
微卫星DNA又称简单重复序列 (simplesequence repeat,SSR),是真核生物基因组中以少数几个核苷酸 (一般为2~6个)为单位的串联重复序列[3]。微卫星DNA标记具有稳定可靠、多态性高的优点,已被广泛应用于水产动物种质资源评估[4-5]、种群遗传结构分析[6-7]、遗传图谱构建[8-9]及系统进化分析[10-11]等研究中。章芸等[12]利用8个微卫星标记对湖北荆州、浙江海宁等7个乌龟养殖群体进行了遗传多样性和遗传结构分析,发现这7个养殖群体均表现出较高的多态性且群体间等位基因扩散受到一定程度的限制。Bu等[13]通过微卫星分子标记对广州、南宁和芜湖等5个乌龟群体遗传结构分析表明,这几个群体存在着较大的遗传分化。罗相忠等[14]采用微卫星分析了鲢(Hypophthalmichthys molitrix)和长丰鲢世代间的遗传多样性和遗传结构,结果表明,经过连续3代利用,长丰鲢的遗传结构发生了改变,遗传多样性呈下降趋势,但遗传多样性水平仍较高,这为长丰鲢优良性状的进一步维持提供了依据。因此,微卫星DNA是用来评价水产动物选育效果的良好工具。本研究中,采用微卫星标记检测了乌龟5个选育世代群体的遗传多样性及遗传结构,以期为乌龟优良品种的选育提供基础数据。
1 材料与方法
1.1 材料
试验用乌龟群体为湖北京山乌龟原种及安徽大别山野生乌龟种群体。
1.2 方法
1.2.1 样本采集与DNA提取 以生长速度为选育指标,对两个群体选择体长椭圆形、体色棕黑的乌龟新品种。乌龟选育基础群体建立于2010年,结合安徽蓝田农业集团有限公司工厂化养殖模式,每4年选育产生一代,每代选留率为10%。从5个选育世代(基础群体F0代及F1、F2、F3、F4代)的每一代中随机选取60只,用干净剪刀剪取乌龟指甲,放入无水乙醇中固定后于-20 ℃下保存。
利用MicroElute Genomic DNA Kit(Omega Biotek,Inc.Norcross,Georgia,USA)提取指甲样品DNA。用琼脂糖凝胶电泳检测DNA的纯度及完整性,用NanoQTM微型分光光度计检测DNA浓度。
1.2.2 微卫星引物的筛选 将Ye等[15]报道的8对微卫星引物及本研究中乌龟转录组测序中随机选择的50对微卫星标记进行多态性检验。选择10只乌龟基因组DNA为模板,使用M13通用接头序列(TGTAAAACGACGGCCAGT)加到每对引物的F引物5′方向,合成带不同荧光基团的M13接头序列。
PCR反应体系(10 μL):2×Taq PCR Master Mix 5 μL,10 p mol/L上、下游引物混合物1 μL,基因组DNA(50~200 ng)1 μL,去离子水3 μL。PCR扩增程序:96 ℃下预变性3 min;96 ℃下变性30 s,最适温度下退火30 s,72 ℃下延伸1 min,共进行30个循环;最后在72 ℃下再延伸10 min,在12 ℃条件下保存。
带荧光的PCR产物经DNA测序仪ABI 3730xl进行荧光电泳检测,采用GeneMarker 2.2.0软件对原始数据进行条带分型,最终筛选出12对微卫星引物进行后续分析。引物均由广州天一辉远基因科技有限公司合成。12对微卫星引物的序列、退火温度和产物片段大小等信息见表1。
表1 乌龟多态性微卫星位点信息
Tab.1 Information on polymorphic microsatellite loci of Mauremys reevesii
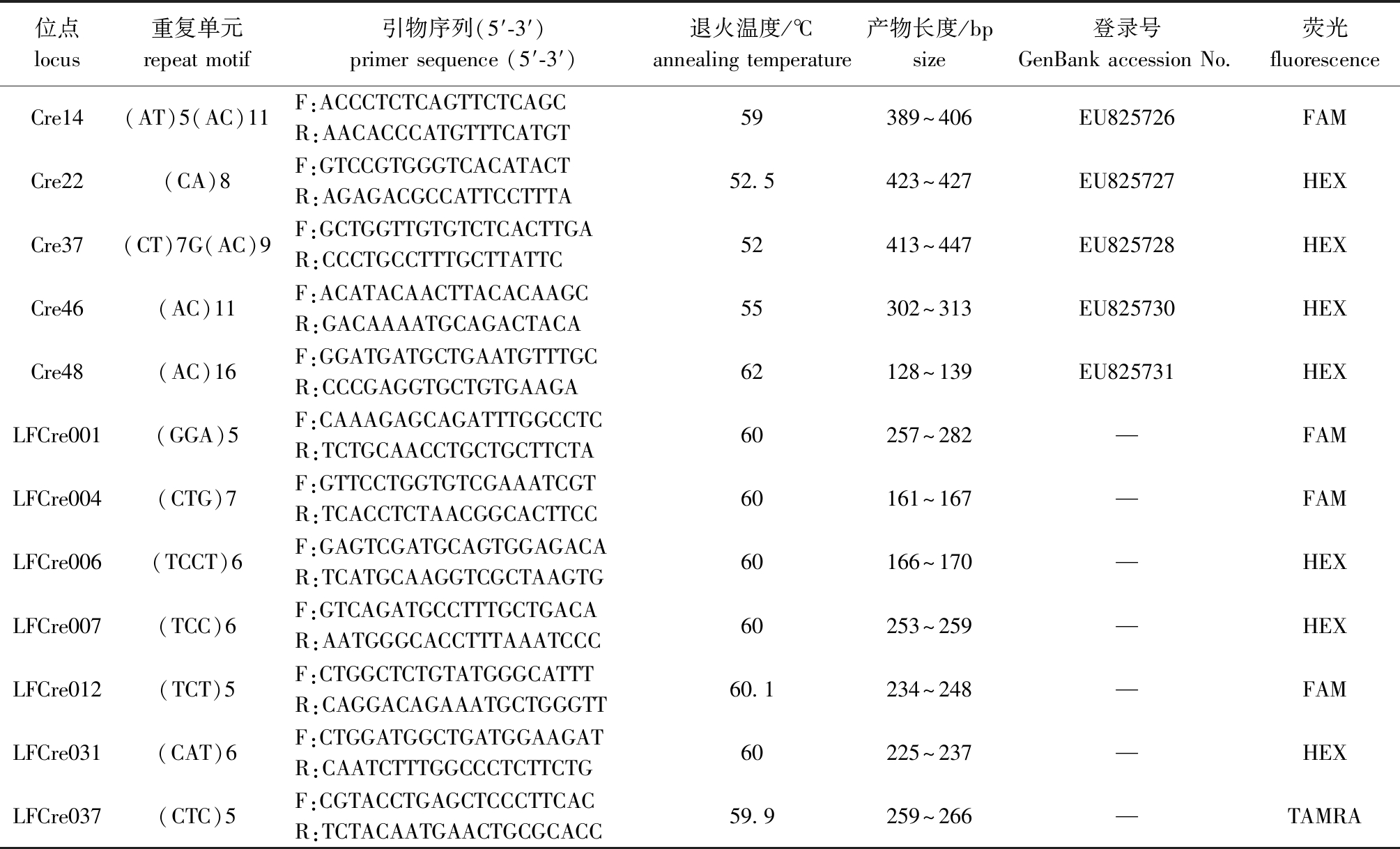
位点locus重复单元repeat motif引物序列(5′-3′)primer sequence (5′-3′)退火温度/℃annealing temperature产物长度/bpsize登录号GenBank accession No.荧光fluorescenceCre14(AT)5(AC)11F:ACCCTCTCAGTTCTCAGCR:AACACCCATGTTTCATGT59389^406EU825726FAMCre22(CA)8F:GTCCGTGGGTCACATACTR:AGAGACGCCATTCCTTTA52.5423^427EU825727HEXCre37(CT)7G(AC)9F:GCTGGTTGTGTCTCACTTGAR:CCCTGCCTTTGCTTATTC52413^447EU825728HEXCre46(AC)11F:ACATACAACTTACACAAGCR:GACAAAATGCAGACTACA55302^313EU825730HEXCre48(AC)16F:GGATGATGCTGAATGTTTGCR:CCCGAGGTGCTGTGAAGA62128^139EU825731HEXLFCre001(GGA)5F:CAAAGAGCAGATTTGGCCTCR:TCTGCAACCTGCTGCTTCTA60257^282—FAMLFCre004(CTG)7F:GTTCCTGGTGTCGAAATCGTR:TCACCTCTAACGGCACTTCC60161^167—FAMLFCre006(TCCT)6F:GAGTCGATGCAGTGGAGACAR:TCATGCAAGGTCGCTAAGTG60166^170—HEXLFCre007(TCC)6F:GTCAGATGCCTTTGCTGACAR:AATGGGCACCTTTAAATCCC60253^259—HEXLFCre012(TCT)5F:CTGGCTCTGTATGGGCATTTR:CAGGACAGAAATGCTGGGTT60.1234^248—FAMLFCre031(CAT)6F:CTGGATGGCTGATGGAAGATR:CAATCTTTGGCCCTCTTCTG60225^237—HEXLFCre037(CTC)5F:CGTACCTGAGCTCCCTTCACR:TCTACAATGAACTGCGCACC59.9259^266—TAMRA
1.2.3 荧光PCR扩增体系构建及产物检测 利用上述筛选好的微卫星引物,对乌龟选育世代300个样本进行PCR扩增,反应体系及PCR扩增程序见“1.2.2节”。
取3 μL荧光PCR产物进行琼脂糖凝胶电泳鉴定,检测PCR条带是否单一、产物片段大小是否与预期一致。条带单一且大小相符的产物,用对照DNA Marker的浓度进行定量,将所有产物稀释至相同的浓度范围,然后利用DNA测序仪ABI 3730xl进行毛细管电泳检测。
1.3 数据处理
通过GenAlEx 6.503软件[16]计算各位点的等位基因数(number of alleles,Na)、有效等位基因数(number of effective alleles,Ne)、平均观测杂合度(observed heterozygosity,Ho)、平均期望杂合度(expected heterozygosity,He)、固定指数F及两两群体间遗传分化系数Fst等遗传参数,采用Cervus 3.0.7软件[17]计算多态信息含量(polymorphism information content,PIC)。利用Genepop 4.3软件[18]和Nei[19]的方法计算选育世代间遗传距离D和遗传相似性系数I。通过UPGMA 法构建群体的系统进化树,分析群体间的亲缘关系。
2 结果与分析
2.1 微卫星引物筛选及遗传多态性
从表2可见:12个微卫星位点在300个乌龟样品中共检测出103个等位基因,平均等位基因数8.583,其中,Cre37和Cre48等位基因数目较多,分别为19和20个,Cre14次之,为10个;有效等位基因数为1.209~12.390,平均值为3.632;多态信息含量为 0.165~0.914,平均值为0.541;期望杂合度为0.173~0.919,平均值为0.581;观测杂合度为 0.054~0.862,平均值为0.479;近交系数为-0.061~0.863。其中,Cre14、Cre22、Cre37、Cre48、LFCre007、LFCre012和LFCre031(1.000>PIC>0.500)为高度多态位点,Cre46、LFCre001、LFCre006和LFCre037(0.500>PIC>0.250)属于中度多态位点。
表2 乌龟微卫星 DNA 位点的特征
Tab.2 Characterization of the microsatellite loci isolated from Mauremys reevesii
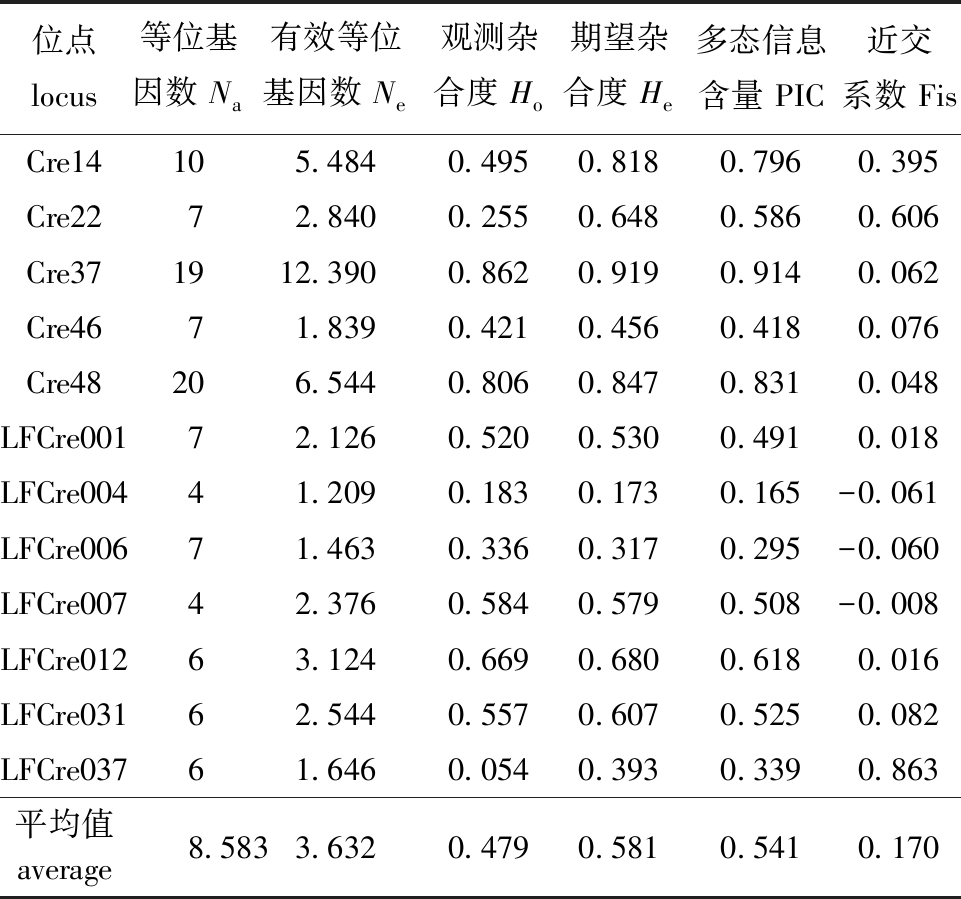
位点 locus等位基因数 Na有效等位基因数 Ne观测杂合度 Ho期望杂合度 He多态信息含量 PIC近交系数 FisCre14105.4840.4950.8180.7960.395Cre2272.8400.2550.6480.5860.606Cre371912.3900.8620.9190.9140.062Cre4671.8390.4210.4560.4180.076Cre48206.5440.8060.8470.8310.048LFCre00172.1260.5200.5300.4910.018LFCre00441.2090.1830.1730.165-0.061LFCre00671.4630.3360.3170.295-0.060LFCre00742.3760.5840.5790.508-0.008LFCre01263.1240.6690.6800.6180.016LFCre03162.5440.5570.6070.5250.082LFCre03761.6460.0540.3930.3390.863平均值average8.5833.6320.4790.5810.5410.170
2.2 乌龟选育世代遗传多样性
从表3可见:12个乌龟微卫星位点在5个选育世代群体中Na和Ne值最高的位点均为Cre37,其次是位点Cre48;5个选育世代群体中He值最高的位点均为Cre37,F0、F1、F2和F3代Ho值最高的位点为Cre37,F4代群体Ho值最高的位点为Cre48。
表3 12个微卫星位点在乌龟选育世代的遗传学特征
Tab.3 Genetic characterizations of 12 polymorphism SSR loci in Mauremys reevesii
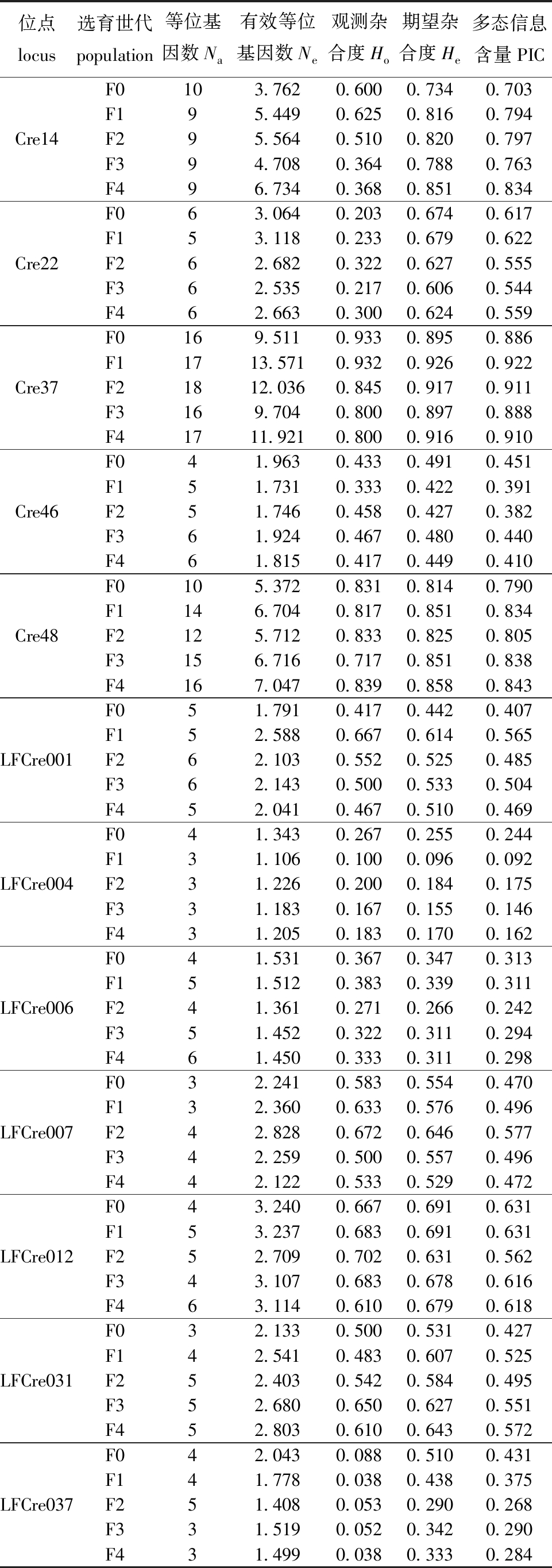
位点 locus选育世代population等位基因数Na有效等位基因数Ne观测杂合度Ho期望杂合度He多态信息含量PICF0103.7620.6000.7340.703F195.4490.6250.8160.794Cre14F295.5640.5100.8200.797F394.7080.3640.7880.763F496.7340.3680.8510.834F063.0640.2030.6740.617F153.1180.2330.6790.622Cre22F262.6820.3220.6270.555F362.5350.2170.6060.544F462.6630.3000.6240.559F0169.5110.9330.8950.886F11713.5710.9320.9260.922Cre37F21812.0360.8450.9170.911F3169.7040.8000.8970.888F41711.9210.8000.9160.910F041.9630.4330.4910.451F151.7310.3330.4220.391Cre46F251.7460.4580.4270.382F361.9240.4670.4800.440F461.8150.4170.4490.410F0105.3720.8310.8140.790F1146.7040.8170.8510.834Cre48F2125.7120.8330.8250.805F3156.7160.7170.8510.838F4167.0470.8390.8580.843F051.7910.4170.4420.407F152.5880.6670.6140.565LFCre001F262.1030.5520.5250.485F362.1430.5000.5330.504F452.0410.4670.5100.469F041.3430.2670.2550.244F131.1060.1000.0960.092LFCre004F231.2260.2000.1840.175F331.1830.1670.1550.146F431.2050.1830.1700.162F041.5310.3670.3470.313F151.5120.3830.3390.311LFCre006F241.3610.2710.2660.242F351.4520.3220.3110.294F461.4500.3330.3110.298F032.2410.5830.5540.470F132.3600.6330.5760.496LFCre007F242.8280.6720.6460.577F342.2590.5000.5570.496F442.1220.5330.5290.472F043.2400.6670.6910.631F153.2370.6830.6910.631LFCre012F252.7090.7020.6310.562F343.1070.6830.6780.616F463.1140.6100.6790.618F032.1330.5000.5310.427F142.5410.4830.6070.525LFCre031F252.4030.5420.5840.495F352.6800.6500.6270.551F452.8030.6100.6430.572F042.0430.0880.5100.431F141.7780.0380.4380.375LFCre037F251.4080.0530.2900.268F331.5190.0520.3420.290F431.4990.0380.3330.284
从表4可见,F0、F1、F2、F3及F4代群体的Na平均值为6.700,Ne平均值为3.497,He平均值为0.497,Ho平均值为0.574。
表4 5个乌龟选育世代的遗传多样性
Tab.4 Genetic diversity in 5 selective breeding generations of Mauremys reevesii
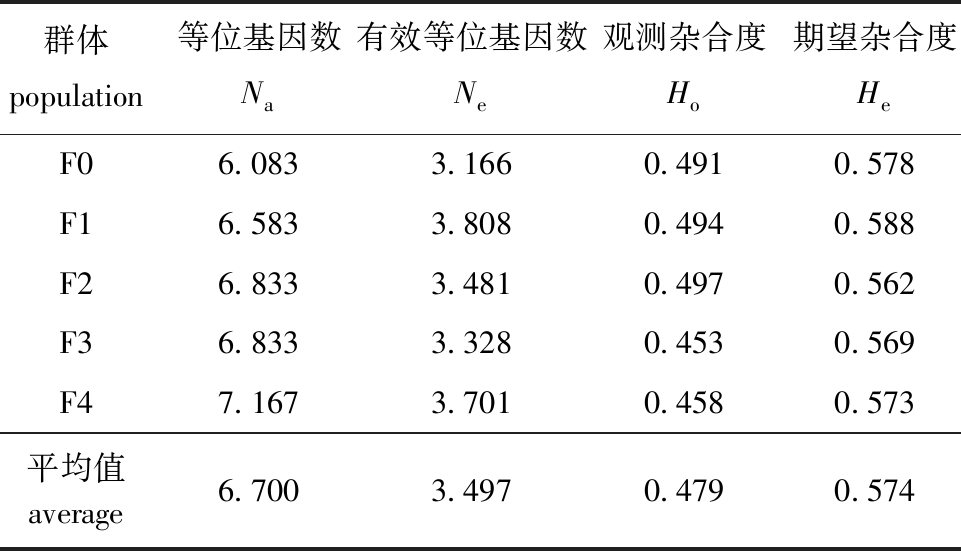
群体population等位基因数Na有效等位基因数Ne观测杂合度Ho期望杂合度HeF06.0833.1660.4910.578F16.5833.8080.4940.588F26.8333.4810.4970.562F36.8333.3280.4530.569F47.1673.7010.4580.573平均值 average6.7003.4970.4790.574
2.3 乌龟选育群体间遗传结构
乌龟5个选育世代群体之间的Fst值为0.004~0.012,小于0.05,说明乌龟选育世代群体间的遗传分化较小(表5)。分子方差(AMOVA)分析显示,0%的遗传变异存在于选育群体之间,100%的遗传变异存在于群体的个体内,个体的变异是乌龟选育世代变异的主要来源(表6)。计算得出5个选育世代的Nei氏遗传距离仅为0.021~0.037(表7)。依据5个选育世代群体间的遗传距离值,采用UPGMA法构建聚类图,结果显示,乌龟5个选育世代聚为2个大的类群,F1代群体与F2代群体汇成一支,与F3代群体和F4代群体汇成的一支构成姐妹群,最后与F0代群体聚在一起(图1)。

图1 基于Nei氏无偏遗传距离的UPGMA聚类
Fig.1 UPGMA clustering based on Nei’s unbiased genetic distance
表5 5个乌龟选育世代间遗传分化系数(Fst)及基因流(Nm)
Tab.5 Comparing pairwise values of Fst and Nm among 5 selective breeding generations of Mauremys reevesii
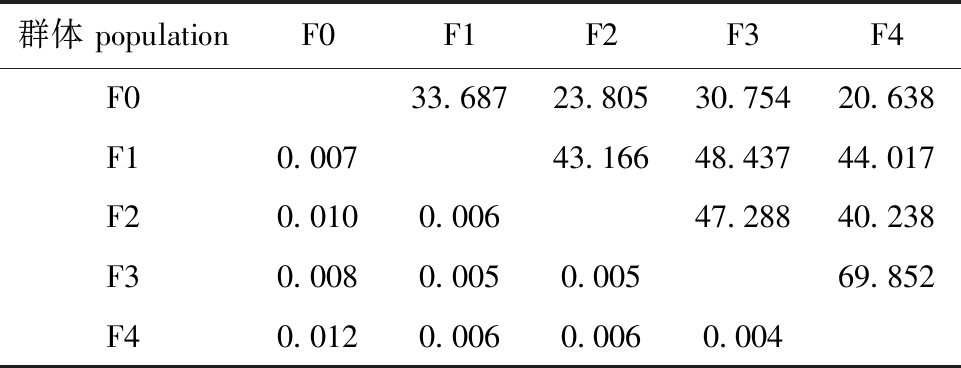
注:对角线以下为遗传分化系数,对角线以上为基因流。
Note:The data below the diagonal represent Fst value,and means above the diagonal represent genetic distance.
群体 populationF0F1F2F3F4F033.68723.80530.75420.638F10.00743.16648.43744.017F20.0100.00647.28840.238F30.0080.0050.00569.852F40.0120.0060.0060.004
表6 5个乌龟选育世代的分子变异方差分析
Tab.6 Hierarchical AMOVA analysis of 5 selective breeding generations of Mauremys reevesii
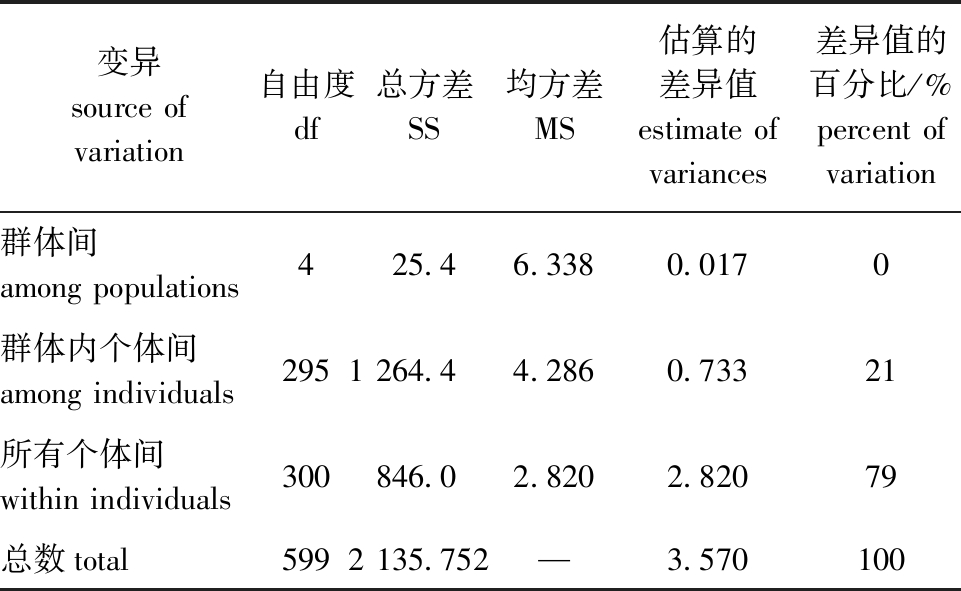
变异source of variation自由度df总方差SS均方差MS估算的差异值estimate of variances差异值的百分比/%percent of variation群体间 among populations425.46.3380.0170群体内个体间 among individuals2951 264.44.2860.73321所有个体间 within individuals300846.02.8202.82079总数total5992 135.752—3.570100
表7 5个乌龟选育世代的Nei氏遗传距离
Tab.7 Nei’s standard genetic distance among 5 selective breeding generations of Mauremys reevesii
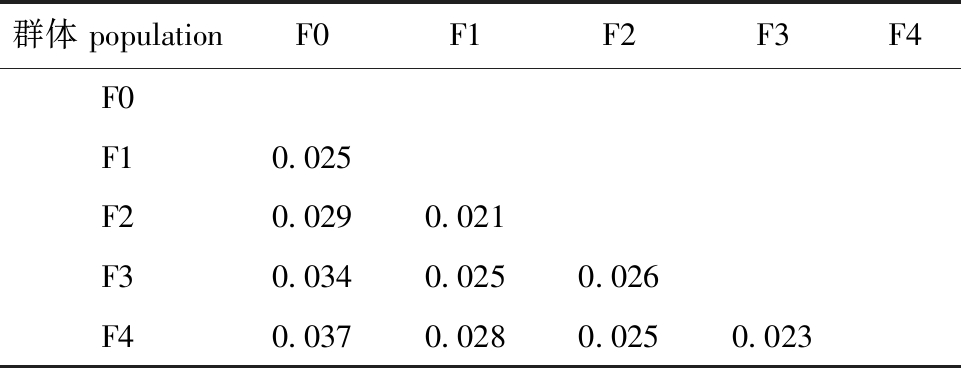
群体 populationF0F1F2F3F4F0F10.025F20.0290.021F30.0340.0250.026F40.0370.0280.0250.023
3 讨论
3.1 微卫星标记的多态性及遗传学特征
微卫星标记因具有稳定性好、多态性高和便于检测等优点,已在水产动物品种选育、种质鉴定和物种遗传多样性分析等研究中广泛应用[20-21]。多态信息含量是度量一个遗传标记多态性所含信息量的指标,反映一个后代所获得的某个等位基因标记来自其亲代同一个等位基因标记的可能性[22],为遗传多样性提供了初步结论。当PIC>0.5时,属于高度多态性位点;当0.25≤PIC≤0.5时,属于为中度多态位点;当PIC<0.25时,属于低度多态位点[23]。本研究中,利用微卫星标记对乌龟5个选育世代群体的遗传多样性及遗传结构进行了检测,在筛选的12个微卫星位点中,共检测到等位基因数103个,平均等位基因数Na为8.583;12个位点多态信息含量平均值为0.541,其中11个位点均属于中度或高度多态位点(表2),表明选择的微卫星位点可作为乌龟选育世代遗传多样性及遗传结构分析的良好评价工具。
3.2 乌龟选育世代的遗传多样性
生物群体的遗传多样性是生态系统多样性和物种多样性的基础和核心。一个生物群体遗传多样性在存在时间上是延续不断的,是进化的基本单位。遗传多样性变异越丰富,群体对环境变化的适应能力越强,进化潜力越大[24-25]。遗传多样性的研究可以揭示该物种的进化历史和潜力。期望杂合度是评价群体生物遗传多样性的最适参数[26]。Nei[19]也指出,期望杂合度(Nei氏基因多样度)是度量种群基因多样性程度的优良指标。本研究中,乌龟选育世代F0、F1、F2、F3及F4代群体中每个微卫星位点Na为3~18个(表3),平均Ho为0.479,平均He为0.547,总体上群体的基因丰富度较高(表4)。5个乌龟选育世代群体F3、F4代群体的平均Ho要小于F0、F1、F2代群体(表4),可见随着选育的进行,乌龟5个世代的平均观测杂合度有所降低,但差距不大。龟鳖动物相对鱼类等水产动物繁殖力较低,每年产卵3~4窝,每窝5~16枚龟卵。因此,随着选育的进行,乌龟各世代的平均观测杂合度相差不大的现象,可能与龟鳖动物低繁殖力的生物学特性相关,这与对中华鳖(Pelodiscus sinensis)选育研究结果类似[27]。本研究中,5个选育世代群体中期望杂合度最高的微卫星位点均为Cre37,这与章芸[28]的研究结果类似,说明该微卫星位点作为乌龟选育世代遗传多样性及遗传结构分析的评价工具较为合适。
3.3 乌龟选育世代的遗传变异
遗传分化指数是评价群体间遗传分化程度的重要指标。当Fst值为0~0.05时,表明群体间遗传分化较小,可以不予考虑;当Fst值为0.05~0.15时,表明群体间存在中等程度的分化;当Fst值为0.15~0.25时,表明群体间存在较大的遗传分化;当Fst值为0.25以上时,表明群体间遗传分化显著[29]。本研究中,乌龟5个选育世代间的Fst值为0.004~0.012(表5),且无变异来自群体间,100%的变异均来自群体内部(表6),表明乌龟选育世代间的遗传分化程度较低,这与长丰鲢遗传分化系数在相邻世代之间相继减少的结果相类似[14]。遗传距离是衡量群体间遗传分化程度的重要指标。根据Thorpe[30]提出的理论,同物种群体间的遗传距离为0.03~0.20。本研究中,通过Nei[19]的方法计算得出5个选育世代遗传距离仅为0.021~0.037(表7),表明仅4个人工选育世代过程对乌龟选育群体的遗传结构并未产生较大影响,还需要进一步选育。选择育种往往需要连续多代选育才能获得可稳定遗传的优良性状。但随着选育代数增加,群体遗传多样性往往会不断下降,影响进一步选育的效果。适当拓宽选育群体的遗传基础可为其选育工作的持续进行提供有力保障[31]。由此可见,乌龟选育群体内还保持有一定的遗传多样性,有进一步选育的潜力。
4 结论
1)乌龟5个选育世代的Ho分别为0.491、0.494、0.497、0.458和0.453,表明乌龟选育世代的遗传多样性有所下降,但差异不大。
2)乌龟5个选育世代间的Fst值为0.004~0.012,且100%的变异均来自群体内部,表明乌龟5个选育世代间的遗传分化程度较小,具有进一步选育的潜力。
[1] 杜杰,孙建义,卢亚萍.乌龟的营养价值及营养需要[J].中国饲料,2006(15):32-34.
DU J,SUN J Y,LU Y P.The nutrition value and nutrition requirement of Chinemys reevesii[J].China Feed,2006(15):32-34.(in Chinese)
[2] 万全,彭步旭,徐勇,等.乌龟人工繁殖和稚龟培育方法研究[J].现代农业科技,2010(19):294-295,309.
WAN Q,PENG B X,XU Y,et al.Study on artificial breeding of turtles and cultivation of young turtles[J].Modern Agricultural Sciences and Technology,2010(19):294-295,309.(in Chinese)
[3] 王伟,尤锋,高天翔,等.鱼类微卫星标记的研究进展[J].海洋科学,2006,30(10):81-86.
WANG W,YOU F,GAO T X,et al.Review on microsatellite markers in fish[J].Marine Sciences,2006,30(10):81-86.(in Chinese)
[4] 闫春梅,张雅斌,郑伟,等.鸭绿江鲤鱼种质资源的微卫星分析[J].东北农业大学学报,2011,42(12):102-106.
YAN C M,ZHANG Y B,ZHENG W,et al.Genetic relationship in wild carps using microsatellite markers[J].Journal of Northeast Agricultural University,2011,42(12):102-106.(in Chinese)
[5] TANG S J,LI S F,CAI W Q,et al.Microsatellite analysis of variation among wild,domesticated,and genetically improved populations of blunt snout bream (Megalobrama amblycephala)[J].Zoological Research,2014,35(2):108-117.
[6] 郭昱嵩,王中铎,谢子强,等.红鳍笛鲷微卫星DNA标记的开发与遗传多样性分析[J].广东海洋大学学报,2011,31(4):13-17.
GUO Y S,WANG Z D,XIE Z Q,et al.Isolation and genetic diversity analysis of microsatellite DNA in Lutjanus erythopterus[J].Journal of Guangdong Ocean University,2011,31(4):13-17.(in Chinese)
[7] CHEN Z Z,HU C X,WANG L,et al.Microsatellite-based genetic structure and differentiation of goldfish (Carassius auratus) with sarcoma[J].Open Journal of Animal Sciences,2015,5(1):36-43.
[8] MCCONNELL S K,BEYNON C,LEAMON J,et al.Microsatellite marker based genetic linkage maps of Oreochromis aureus and O.niloticus(Cichlidae):extensive linkage group segment homologies revealed[J].Animal Genetics,2000,31(3):214-218.
[9] 宋文涛,张潇峮,廖小林,等.牙鲆微卫星标记遗传连锁图谱的构建[J].农业生物技术学报,2011,19(6):981-987.
SONG W T,ZHANG X Q,LIAO X L,et al.Construction of a microsatellite-based genetic linkage map in Japanese flounder(Paralichthys olivaceus)[J].Journal of Agricultural Biotechnology,2011,19(6):981-987.(in Chinese)
[10] 李景芬,夏正龙,栾生,等.五个罗氏沼虾群体遗传多样性的微卫星分析[J].水生生物学报,2020,44(6):1208-1214.
LI J F,XIA Z L,LUAN S,et al.Genetic diversity analysis of five populations of Macrobrachium rosenbergii using microsatellite markers[J].Acta Hydrobiologica Sinica,2020,44(6):1208-1214.(in Chinese)
[11] LEI Y,ZHOU Y,PRICE M,et al.Genome-wide characterization of microsatellite DNA in fishes:survey and analysis of their abundance and frequency in genome-specific regions[J].BMC Genomics,2021,22(1):421.
[12] 章芸,俞丹娜,杜卫国,等.微卫星标记分析乌龟养殖群体的遗传多样性[J].水产学报,2010,34(11):1636-1644.
ZHANG Y,YU D N,DU W G,et al.Microsatellite DNA analysis of genetic diversity among captive breeding stocks of Chinese pond turtle(Chinemys reevesii)[J].Journal of Fisheries of China,2010,34(11):1636-1644.(in Chinese)
[13] BU X,WANG X,LU W,et al.Genetic diversity of the captive Chinese pond turtle (Mauremys reevesii) populations in China assessed by microsatellite markers[J].The Journal of Animal and Plant Sciences,2019, 29(4):1160-1168.
[14] 罗相忠,覃维敏,梁宏伟,等.基于微卫星分析的长丰鲢种质资源遗传监测[J].水生生物学报,2022,46(5):725-734.
LUO X Z,QIN W M,LIANG H W,et al.Genetic monitoring of Changfeng silver carp base on microsatellite[J].Acta Hydrobiologica Sinica,2022,46(5):725-734.(in Chinese)
[15] YE R H,ZHENG R Q,WANG L,et al.Polymorphic microsatellite loci in the Chinese pond turtle (Chinemys reevesii)[J].Conservation Genetics,2009,10(4):1045-1048.
[16] PEAKALL R,SMOUSE P E.Genalex 6:genetic analysis in excel population genetic software for teaching and research[J].Molecular Ecology Notes,2006,6(1):288-295.
[17] KALINOWSKI S T,TAPER M L,MARSHALL T C.Revising how the computer program Cervus accommodates genotyping error increases success in paternity assignment[J].Molecular Ecology,2007,16(5):1099-1106.
[18] ROUSSET F.Genepop’007:a complete re-implementation of the Genepop software for Windows and Linux[J].Molecular Ecology Resources,2008,8(1):103-106.
[19] NEI M.Molecular evolutionary genetics[M].New York:Columbia University Press,1987.
[20] 张新辉,高泽霞,罗伟,等.雌核发育团头鲂的形态和遗传特征分析[J].水生生物学报,2015,39(1):126-132.
ZHANG X H,GAO Z X,LUO W,et al.Studies on morphological characteristics and genetic analysis of the gynogenesis blunt snout bream(Megalobrama amblycephala)[J].Acta Hydrobiologica Sinica,2015,39(1):126-132.(in Chinese)
[21] 王丰,张家华,沈玉帮,等.青鱼野生与养殖群体遗传变异的微卫星分析[J].水生生物学报,2019,43(5):939-944.
WANG F,ZHANG J H,SHEN Y B,et al.Microsatellite analysis of genetic variation of wild and cultural populations in black carp Mylopharyngodon piceus[J].Acta Hydrobiologica Sinica,2019,43(5):939-944.(in Chinese)
[22] 刘丽,刘楚吾.5种笛鲷属鱼类的遗传多样性及分子标记[J].农业生物技术学报,2006,14(3):349-355.
LIU L,LIU C W.Genetic diversity and molecular markers of 5 species of snappers[J].Journal of Agricultural Biotechnology,2006,14(3):349-355.(in Chinese)
[23] BOTSTEIN D,WHITE R L,SKOLNICK M,et al.Construction of a genetic linkage map in man using restriction fragment length polymorphisms[J].American Journal of Human Genetics,1980,32(3):314-331.
[24] 曾珍.松江鲈不同种群间的分子标记和遗传多样性分析[D].上海:上海海洋大学,2013.
ZENG Z.Molecular markers and genetic diversity analysis in populations of Trachidermus fasciatus[D].Shanghai:Shanghai Ocean University,2013.(in Chinese)
[25] 严骏骢,赵金良,李思发,等.鳙中国土著群体与移居群体遗传变异的AFLP分析[J].中国水产科学,2011,18(2):283-289.
YAN J C,ZHAO J L,LI S F,et al.Genetic variation of bighead carp Aristichthys nobilis from Chinese native populations and introduced populations by AFLP[J].Journal of Fishery Sciences of China,2011,18(2):283-289.(in Chinese)
[26] 王耀嵘,杨尉,任席林,等.金钱鱼基因组微卫星分布特征分析及多态性标记开发[J].广东海洋大学学报,2020,40(4):7-14.
WANG Y R,YANG W,REN X L,et al.Distribution patterns of microsatellites and development of polymorphic markers from Scatophagus argus genome[J].Journal of Guangdong Ocean University,2020,40(4):7-14.(in Chinese)
[27] 程珂,陈辰,史燕,等.基于微卫星的中华鳖(Pelodiscus sinensis)选育世代遗传多样性监测[J].基因组学与应用生物学,2018,37(9):3774-3781.
CHENG K,CHEN C,SHI Y,et al.Genetic diversiy monitoring of selected generations of Chinese soft-shelled turtle(Pelodiscus sinensis) based on microsatellites[J].Genomics and Applied Biology,2018,37(9):3774-3781.(in Chinese)
[28] 章芸.两种淡水龟微卫星标记的筛选及乌龟养殖群体遗传多样性分析[D].金华:浙江师范大学,2010.
ZHANG Y.Polymorphic microsatellite loci in two freshwater turtles and genetic diversity among captive breeding stocks of Chinese pond turtle(Chinemys reevesii)[D].Jinhua:Zhejiang Normal University,2010.(in Chinese)
[29] MCCONNELL S,HAMILTON L,MORRIS D,et al.Isolation of salmonid microsatellite loci and their application to the population genetics of Canadian east coast stocks of Atlantic salmon[J].Aquaculture,1995,137(1/2/3/4):19-30.
[30] THORPE J P.The molecular clock hypothesis:biochemical evolution,genetic differentiation and systematics[J].Annual Review of Ecology and Systematics,1982,13:139-168.
[31] 刘志刚,曹建萌,高风英,等.罗非鱼“粤闽1号”母本选育群体世代间遗传差异的微卫星分析[J].大连海洋大学学报,2021,36(1):16-22.
LIU Z G,CAO J M,GAO F Y,et al.Genetic differentiation analysis of maternal selective breeding generations of tilapia “Yuemin No.1” using microsatellites[J].Journal of Dalian Ocean University,2021,36(1):16-22.(in Chinese)